A Modular Approach to Designing Protein Assemblies
02/11/2021
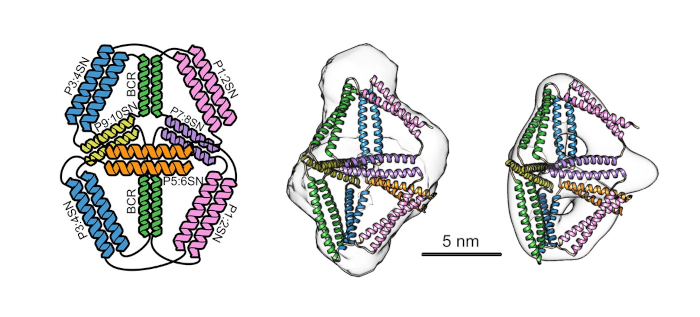
An envisioned protein design becomes reality. At left, scientists designed a protein cage, named BIP18SN, composed of 18 alpha-helical elements. Small-angle X-ray scattering (SAXS; solution X-ray scattering) confirmed that the protein cage in solution adopted the predicted folding conformation (two structures at right). [Reprinted under a Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0/) from Lapenta, et al. 2021. DOI: 10.1038/s41467-021-21184-6.]
Tailor-made nanostructures enable precise control over three-dimensional spatial arrangements and biochemical processes at the molecular level. Biological macromolecules, such as DNA and polypeptides, represent versatile, programmable biomaterials suitable for this purpose.
In designing nanostructures, modularity is a commonly employed concept since it greatly simplifies the design process. In the protein realm, the modular paradigm is achievable by employing α-helical elements, such as coiled-coil (CC) units, as building modules.
Scientists used a type of modular design based on pairwise-interacting CC units, called CC protein origami (CCPO), to create protein cages that self-assembled into oligomers. The conformation adopted by the protein cage in solution was examined using SAXS conducted at the SIBYLS beamline 12.3.1 at the Advanced Light Source. In addition, by introducing a protease cleavage site the researchers created a proteolysis-mediated conformational switch, demonstrating that polyhedral protein cages can be designed to transition between two structural states in response to an external cue.
Self-assembly of CC-based nanostructures from several chains, similarly as in DNA nanotechnology, could facilitate the design of more complex protein assemblies and functionalities.
Related Links
References
Lapenta, F., Aupič, J., Vezzoli, M., et al. 2021. “Self-assembly and regulation of protein cages from pre-organised coiled-coil modules.” Nat Commun 12, 939. [DOI: 10.1038/s41467-021-21184-6]
