Techniques Overview

Structural biology techniques address sample diversity. Critical structures and functions in biology occur across a wide range of distances (subnanometers to centimeters) and times (subpicoseconds to minutes). The DOE Biological and Environmental Research Program makes available a variety of structural biology techniques suited to investigating different sample lengths and experimental time scales. [See bottom of page for image credits.]
At these facilities, BER supports beamlines and resources to enable multiscale structural studies. New insights linking molecular properties to system-level functions can be achieved by integrating these different capabilities to make cross-scale connections and leveraging data from complementary techniques such as super-resolution optical and magnetic resonance imaging.
Cryo-Electron Microscopy and Tomography
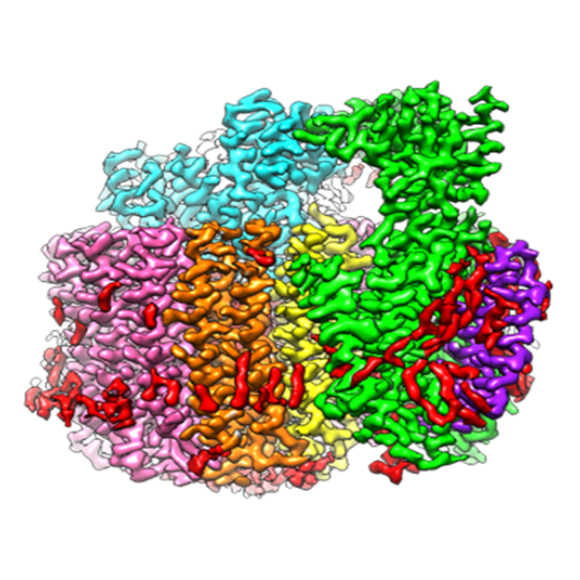
Cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) techniques use electrons to provide images of biological materials frozen in their native states. Materials range from proteins and nucleic acids to very large biological assemblies and complexes, and image resolutions range from nanometer to atomic scales.
- Cryo-EM/ET technique details
- Resources offering this technique:
- Additional enabling capabilities:
Neutron Imaging
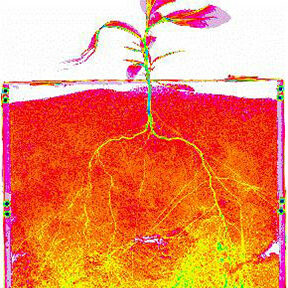
Neutron imaging takes advantage of hydrogen/deuterium contrast and the nondestructive, high-penetrating power of neutrons to study structures in a wide range of hierarchical and complex materials of biological relevance. Applications include studying plant-plant and plant-fungal interactions, soil pore structure and voids under environmentally relevant conditions, fluid transport and interactions in porous media such as the rhizosphere, and cavitation and gas embolism in plant-soil-groundwater systems.
- Neutron Imaging technique details
- Resources offering this technique:
Neutron Macromolecular Crystallography
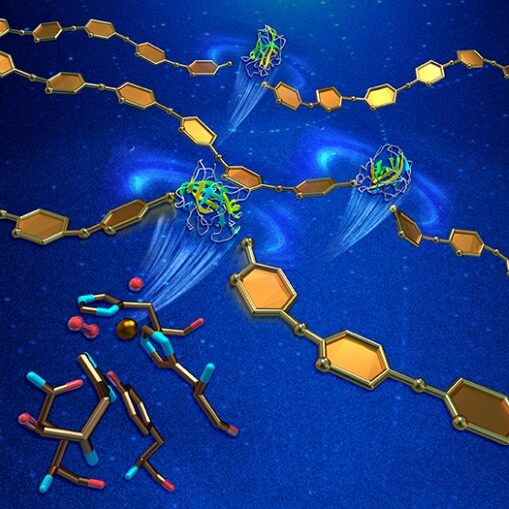
Neutron Macromolecular Crystallography (NMC) provides information about the location of critical hydrogen atoms in protein crystals at atomic resolution. This feature of NMC enables studies of hydrogen bonding networks and the protonation states of catalytic residues.
- NMC technique details
- Resources offering this technique:
Neutron Spectroscopy
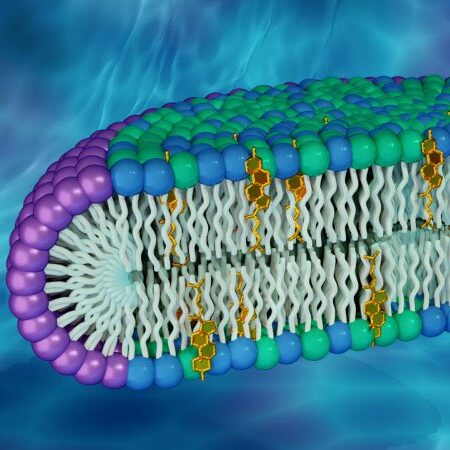
This two-dimensional technique provides information about atomic motions in both time and space. Inelastic and quasi-elastic neutron scattering provide information about vibrational modes, molecular motions, and diffusive properties of biomolecules and their hydration water on the picosecond to nanosecond timescale. Slower motions on the microsecond to millisecond timescale, such as those associated with undulating membranes and domain motions in proteins, are probed by neutron spin echo spectroscopy.
- Neutron Spectroscopy technique details (page coming soon)
- Resources offering this technique:
Small-Angle Neutron Scattering

Like small-angle X-ray scattering (SAXS), small-angle neutron scattering (SANS) is used to study ensemble structures of biological materials with a wide range of length scales in any morphology. SANS, however, can take advantage of the very different neutron scattering cross-sections of hydrogen and deuterium, enabling users to selectively highlight different components within a complex system.
- SANS technique details
- Resources offering this technique:
Soft X-Ray Tomography
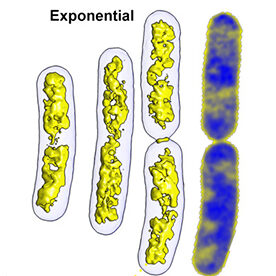
Soft X-ray tomography (SXT) is a non-invasive, three-dimensional imaging technique that can measure volumes, surfaces, interfaces, membranes, and organelle connectivity within intact cells.
- SXT technique details
- Resources offering this technique:
Solution X-Ray Scattering (SAXS)
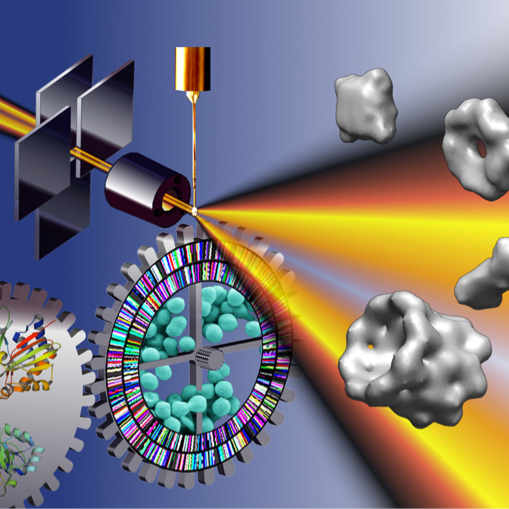
Solution X-ray scattering, often called small-angle X-ray scattering (SAXS), provides structural information about any solution sample. Most investigators use SAXS to characterize the behavior of macromolecules in solution at the nanometer scale. It is an ideal assessment tool in the iterative design cycle of macromolecular engineering and can be used to characterize the degree of ordering in heterogeneous materials such as degrading cellulose.
- Solution X-Ray Scattering technique details
- Resources offering this technique:
Synchrotron Infrared Hyperspectral Imaging
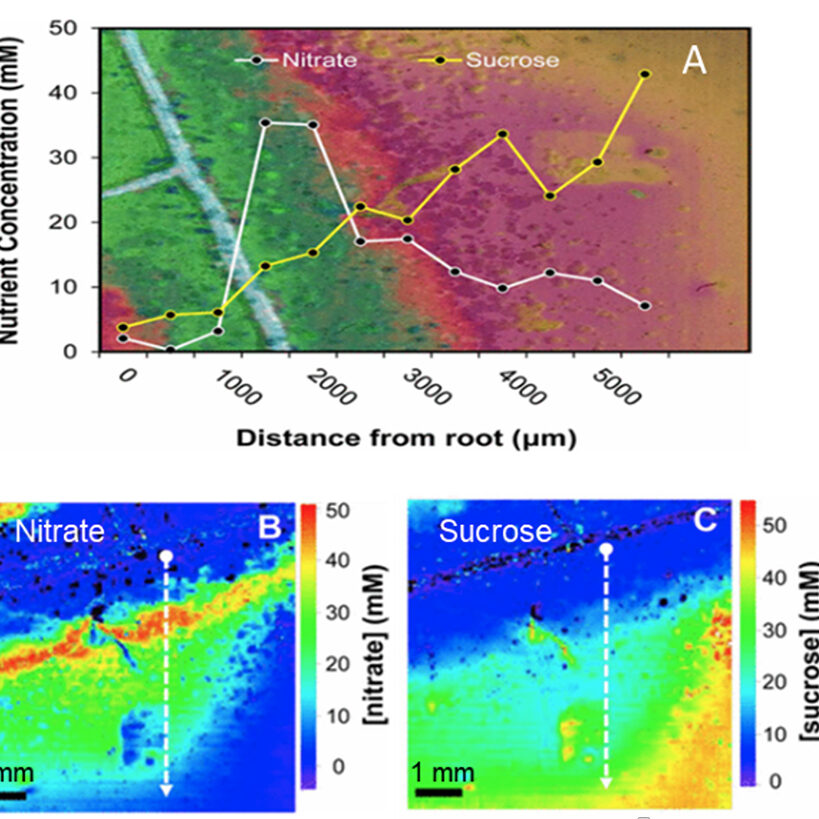
A widely used method of synchrotron infrared hyperspectral imaging is synchrotron-based Fourier-transform infrared imaging (sFTIR), or sFTIR spectromicroscopy. This technique correlates infrared maps with visible microscopy images to map the distributions of molecular compositions of interest within biological samples and to reveal sample morphology and structure.
- Synchrotron Infrared Hyperspectral Imaging technique details
- Resources offering this technique:
X-Ray Absorption and Emission Spectroscopy
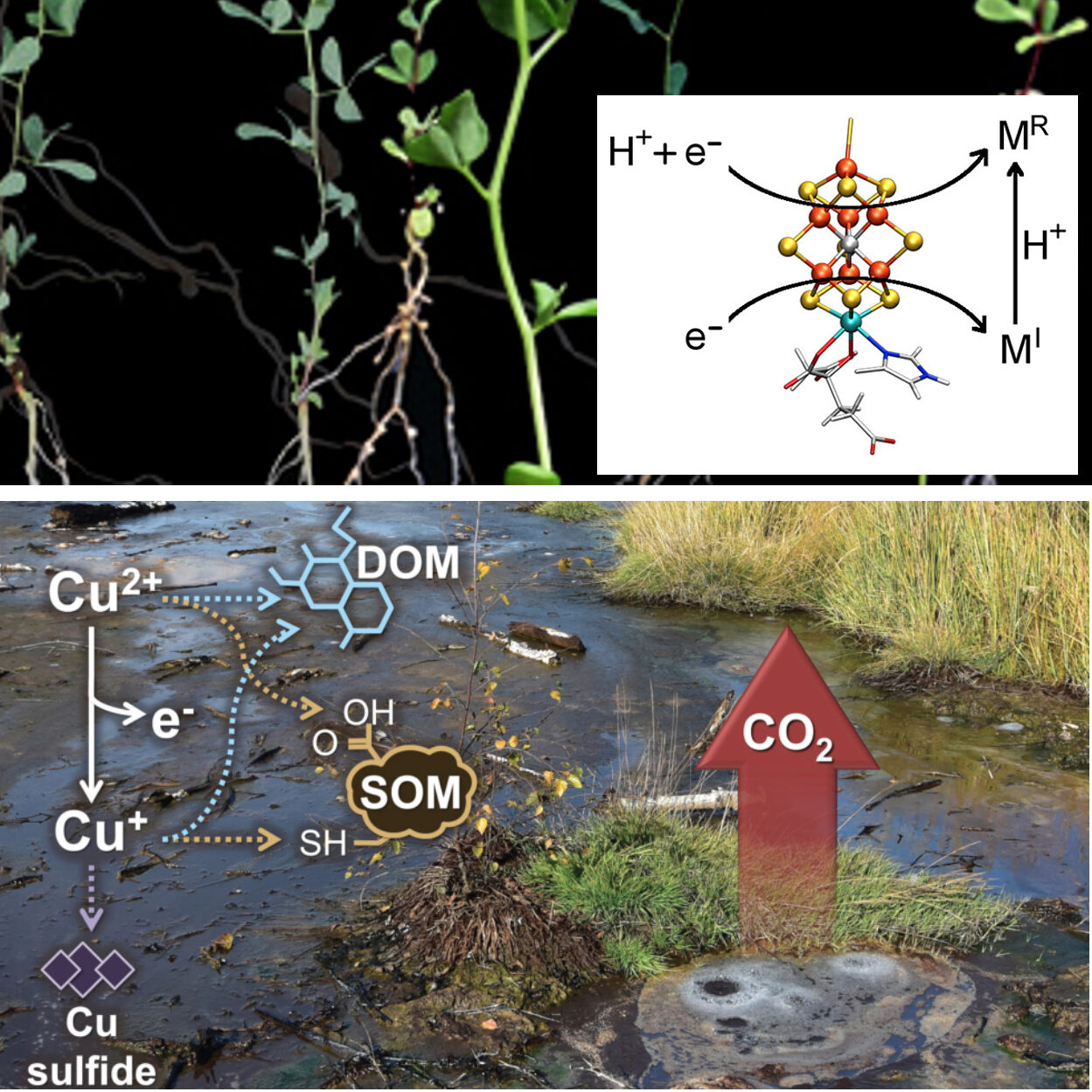
Synchrotron-based X-ray spectroscopy techniques provide a powerful and synergistic toolkit to explore metal interactions within biological and biogeochemical systems and with the environment.
- X-Ray Absorption and Emission Spectroscopy technique details
- BER resources offering this technique:
X-Ray Fluorescence Imaging
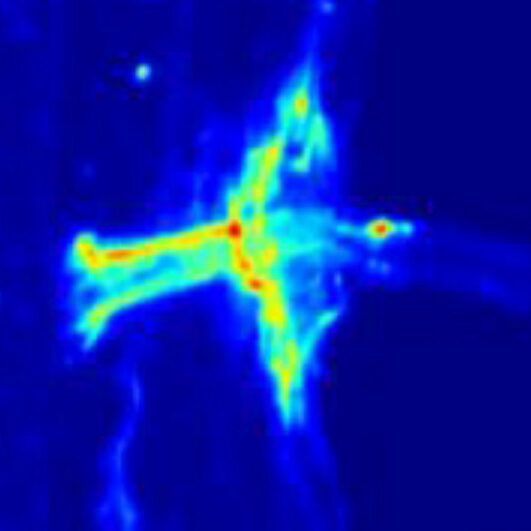
X-ray fluorescence imaging (XRF), or x-ray spectromicroscopy, maps the distributions of elements and chemical species of interest within biological samples. Synchrotron XRF (SXRF) can provide detailed images of element speciation to a resolution of 0.5 µm per pixel, a sensitivity beyond desktop XRF, electron microprobe, or other elemental imaging techniques.
- X-ray Fluorescence Imaging technique details
- Resources offering this technique:
X-ray Footprinting

X-ray footprinting (XFP) is an approach to identifying solvent-accessible regions of a macromolecule (protein or nucleic acid). Solvent accessibility can be an indicator of a binding surface within the macromolecule or an area of conformational movement. When used in combination with other structural biology techniques, footprinting provides complementary or validating information particularly about dynamical motion.
- X-ray Footprinting technique details
- Resources offering this technique:
- X-ray Footprinting of Biological Materials at NSLS-II, BNL (17-BM XFP)
- X-ray Footprinting at the ALS at ALS, LBNL (3.3.1)
X-Ray Macromolecular Crystallography
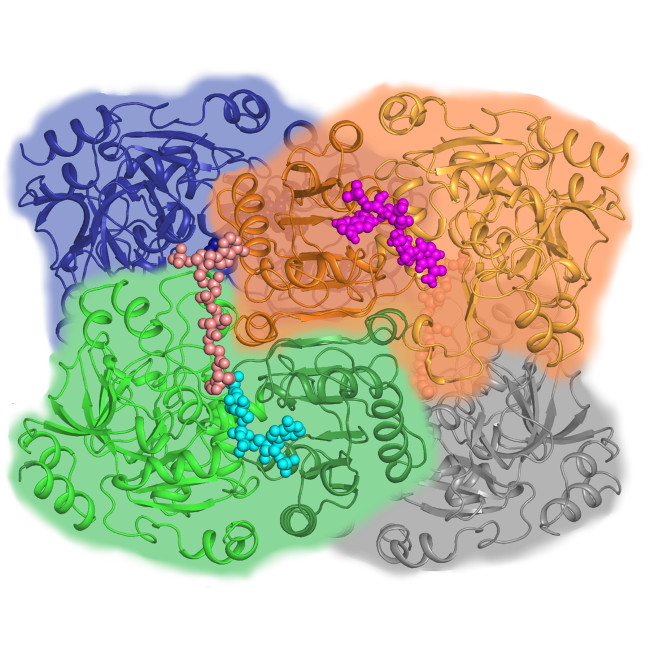
X-ray macromolecular crystallography (MX) uses X-rays to determine the atomic-level structures of biological molecules across a broad range of sample sizes and complexities, including soluble and membrane-bound proteins, enzymes, nucleic acids (RNA and DNA), glycans, other post-translational protein modifications, and complexes of these molecules. MX can determine the structures of complex macromolecular systems and even whole viruses and can investigate bound metabolites or potential drugs, metal ions, and other cofactors.
- X-Ray Macromolecular Crystallography technique details
- Resources offering this technique:
Structural Biology Techniques and Length Scales: Courtesy Stanford Synchrotron Radiation Lightsource at SLAC National Accelerator Laboratory. Individual images left to right: (1) Van Stappen, C., et al. 2019. “Spectroscopic Description of the E1 State of Mo Nitrogenase Based on Mo and Fe X-Ray Absorption and Mössbauer Studies,” Inorganic Chemistry 58(18), 12365–376. DOI:10.1021/acs.inorgchem.9b01951. Reprinted under a Creative Commons License (CC BY 4.0). (2) Kim, Y., et al. 2021. “Tipiracil Binds to Uridine Site and Inhibits Nsp15 Endoribonuclease NendoU from SARS-CoV-2,” Communications Biology 4, 193. DOI:10.1038/s42003-021-01735-9. Reprinted under a Creative Commons License (CC BY 4.0). (3) PDB ID: 6MOR. Roh, S. H., et al. 2020. “Cryo-EM and MD Infer Water-Mediated Proton Transport and Autoinhibition Mechanisms of Vo Complex,” Science Advances 6(41), eabb9605. DOI:10.1126/sciadv.abb9605. (4) Courtesy Thomas SpleIstoesser, www.scistyle.com. See also Vandavasi, V. G., et al. 2016. “A Structural Study of CESA1 Catalytic Domain of Arabidopsis Cellulose Synthesis Complex: Evidence for CESA Trimers,” Plant Physiology 170(1), 123–35. DOI:0.1104/pp.15.01356. (5) Reprinted with permission from Roth, M. S., et al. 2017. “Chromosome-Level Genome Assembly and Transcriptome of the Green Alga Chromochloris zofingiensis Illuminates Astaxanthin Production,” Proceedings of the National Academy of Sciences USA 114(21), E4296–4305. DOI:10.1073/pnas.1619928114. (6) Martin, M. C., et al. 2013. “3D Spectral Imaging with Synchrotron Fourier Transform Infrared Spectro-Microtomography,” Nature Methods 10, 861–64. DOI:10.1038/nmeth.2596. (7) Seyfferth, A. L, et al. 2017. “Evidence for the Root-Uptake of Arsenite at Lateral Root Junctions and Root Apices in Rice (Oryza sativa L.),” Soils 1(1), 3. DOI:10.3390/soils1010003. Reprinted under a Creative Commons License (CC BY 4.0). (8) Courtesy Oak Ridge National Laboratory. See also Dhiman, I., et al. 2017. “Quantifying Root Water Extraction After Drought Recovery Using sub-mm In Situ Empirical Data,” Plant Soil 417, 1–17. DOI:10.1007/s11104-017-3408-5.