Molecular Structure
The following are imaging and characterization techniques that can be used to study molecular structure.
Cryo-Electron Microscopy and Tomography
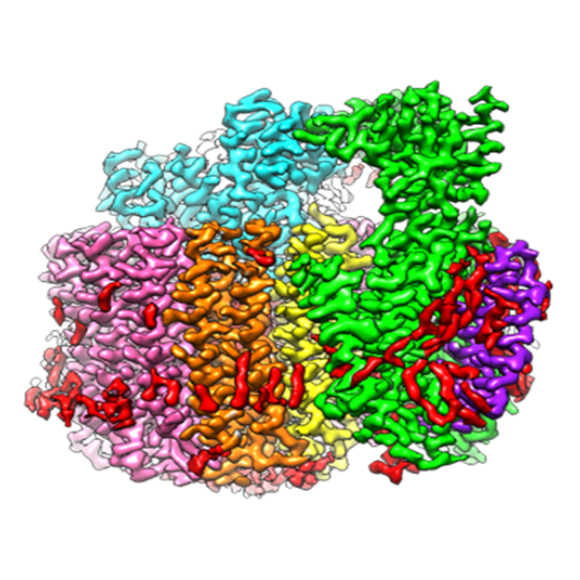
Cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) techniques use electrons to provide images of biological materials frozen in their native states. Materials range from proteins and nucleic acids to very large biological assemblies and complexes, and image resolutions range from nanometer to atomic scales.
- Cryo-EM/ET technique details
- Resources offering this technique:
- Additional enabling capabilities:
Neutron Macromolecular Crystallography
Neutron Macromolecular Crystallography (NMC) provides information about the location of critical hydrogen atoms in protein crystals at atomic resolution. This feature of NMC enables studies of hydrogen bonding networks and the protonation states of catalytic residues.
- NMC technique details
- Resources offering this technique:
Small-Angle Neutron Scattering

Like small-angle X-ray scattering (SAXS), small-angle neutron scattering (SANS) is used to study ensemble structures of biological materials with a wide range of length scales in any morphology. SANS, however, can take advantage of the very different neutron scattering cross-sections of hydrogen and deuterium, enabling users to selectively highlight different components within a complex system.
- SANS technique details
- Resources offering this technique:
Solution X-Ray Scattering (SAXS)
Solution X-ray scattering, often called small-angle X-ray scattering (SAXS), provides structural information about any solution sample. Most investigators use SAXS to characterize the behavior of macromolecules in solution at the nanometer scale. It is an ideal assessment tool in the iterative design cycle of macromolecular engineering and can be used to characterize the degree of ordering in heterogeneous materials such as degrading cellulose.
- Solution X-Ray Scattering technique details
- Resources offering this technique:
X-ray Footprinting

X-ray footprinting (XFP) is an approach to identifying solvent-accessible regions of a macromolecule (protein or nucleic acid). Solvent accessibility can be an indicator of a binding surface within the macromolecule or an area of conformational movement. When used in combination with other structural biology techniques, footprinting provides complementary or validating information particularly about dynamical motion.
- X-ray Footprinting technique details
- Resources offering this technique:
- X-ray Footprinting of Biological Materials at NSLS-II, BNL (17-BM XFP)
- X-ray Footprinting at the ALS at ALS, LBNL (3.3.1)
X-Ray Macromolecular Crystallography
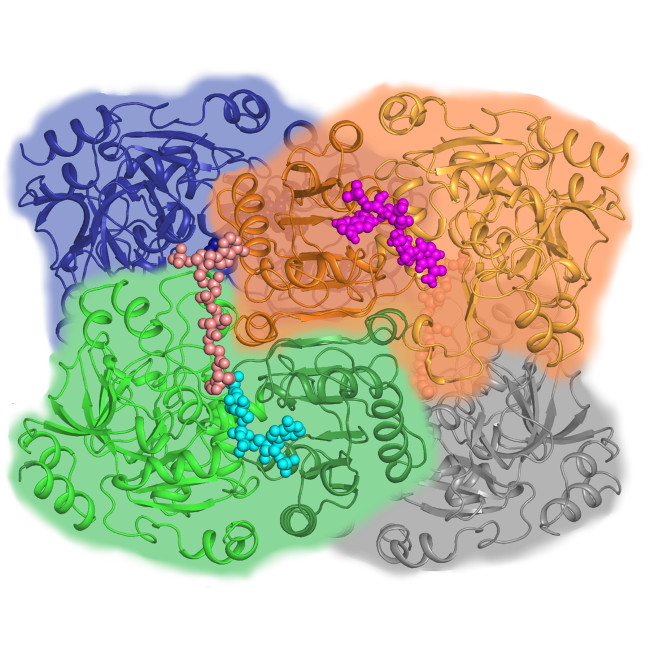
X-ray macromolecular crystallography (MX) uses X-rays to determine the atomic-level structures of biological molecules across a broad range of sample sizes and complexities, including soluble and membrane-bound proteins, enzymes, nucleic acids (RNA and DNA), glycans, other post-translational protein modifications, and complexes of these molecules. MX can determine the structures of complex macromolecular systems and even whole viruses and can investigate bound metabolites or potential drugs, metal ions, and other cofactors.
- X-Ray Macromolecular Crystallography technique details
- Resources offering this technique: