Structural Analysis of Neglected Anaerobic Fungal Enzymes
08/07/2023
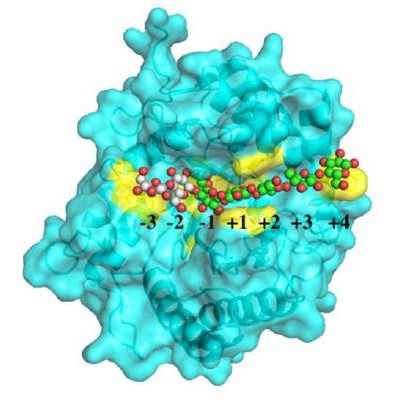
X-ray macromolecular crystallography helps reveal the structure and understand the mechanism of the Piromyces finnis CelD cellulase enzyme (catalytic site is highlighted yellow) bound to an oligosaccharide (red, white, and green spheres).
[Reprinted under a Creative Commons license (CC BY) from Dementiev et al. 2023. DOI:10.1007/s00253-023-12684-0]
The Science
Microbial degradation of lignocellulose is a fundamental biological process crucial to nutrient cycling in nature. The process turns over the equivalent energy of an estimated 640 billion barrels of oil per year. Various microbial enzymes work synergistically to break down hemicellulose, cellulose, and lignin in plant cell walls, with carbohydrate-active enzymes (CAZymes) of the glycoside hydrolase (GH) family largely responsible for cellulose saccharification.
Anaerobic fungi frequently demonstrate cellulolytic activity and hemicellulose degrading power that equals or exceeds the highest performing aerobic microbes and enzyme cocktails. The genomes of these prolific biomass degraders, often found in the guts of large herbivores, harbor a potential wealth of industrially relevant CAZymes.
However, very few have been structurally or biochemically characterized, so very little is known about the characteristics that render the enzymes so efficient. Such knowledge gaps in the biochemistry of individual anaerobic fungal CAZymes themselves, and of fungal cellulosomes which comprise many CAZymes, present a challenge towards designing lignocellulolytic enzyme cocktails that leverage the degradative machinery of anaerobic fungi into useful biotechnologies.
Scientists from Argonne National Laboratory, the University of California-Santa Barbara, and colleagues from three other institutions used X-ray macromolecular crystallography and additional techniques to begin filling the knowledge gap by determining the structure and kinetic rate parameters for a GH family 5 subfamily 4 enzyme, called CelD, from the unique anaerobic fungus Piromyces finnis isolated from the guts of large herbivorous mammals. Notably, CelD’s kinetics do not change with domain fusion, which suggests it is highly modular.
The Impact
The atomic-resolution structure and detailed biochemical characterization of CelD add to a growing understanding of how anaerobic fungal cellulosomes rapidly degrade biomass. Mining anaerobic fungal genomes for better lignocellulolytic enzymes than traditional aerobic microbes may yield higher performing enzymes for industrial biomass valorization applications.
Characterizing the atomic-resolution structure and kinetic properties of the P. finnis CelD GH5 endoglucanase provides additional insight towards gaining biochemical understanding of anaerobic fungal enzyme systems.
The Summary
The study presents the crystal structure of a CelD GH5 catalytic domain in complex with cellotriose, a carbohydrate substrate. CelD is a cellulase that acts strictly as an endo-β-1,4-glucanase with confirmed activity against carboxymethylcellulose, mixed linkage glucan, and xyloglucan.
Observing structural changes during cellotriose substrate binding by the enzyme and linking these structural transformations with the enzyme’s kinetics provided insight into CellD’s catalytic mechanism. The structure presents a platform for rational engineering of this enzyme for higher affinity, thermostability, or activity criteria. The kinetic data indicate that the CelD domains are highly modular and can likely be augmented to be functionalized with other domains that may act synergistically.
Funding
Funding support from the Office of Science (BER), U.S. Department of Energy (DE-SC0010352, DE-SC0020420, and DE-SC0022142), the Institute for Collaborative Biotechnologies through grants W911NF-09-D-0001 and W911NF-19-D-0001 from the U.S. Army Research Office, and the Office of Biological and Environmental Research of the U.S. Department of Energy Joint BioEnergy Institute (JBEI, http:// www. jbei. org) through contract DE-AC02–05CH11231 (Lawrence Berkeley National Laboratory). The Structural Biology Center 19BM beamline is supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Related Links
References
Dementiev, A., et al. 2023. “Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis,” Applied Microbiology and Biotechnology 107, 5999–6011. DOI:10.1007/s00253-023-12684-0
