Cellulose Synthesis Complex
01/12/2016
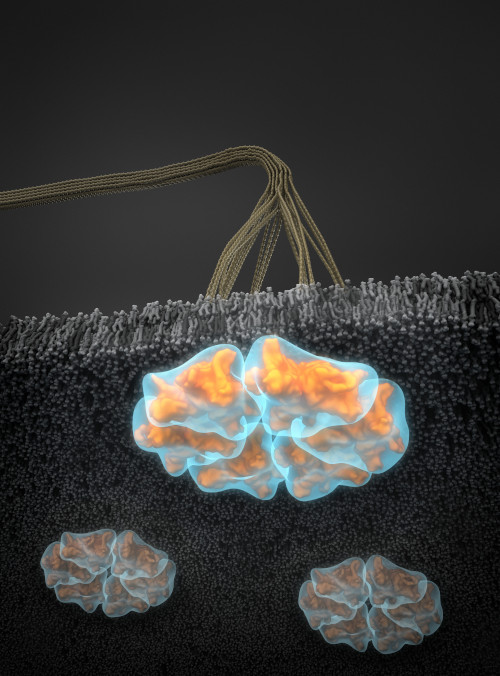
Small-angle scattering data were used to calculate ab initio structures of cellulose synthase (CESA) trimers represented by semi-transparent blue surface envelopes, superposed with computational atomic models in orange. The trimer models are arranged in a hexameric configuration consistent with the rosette shape observed in transmission electron microscope (TEM) images. The view is from the cytosolic side of the membrane. Cellulose microfibrils are visible in the apoplastic space. [Credit: Thomas Splettstoesser, www.scistyle.com, Berlin, Germany. Reprinted from the cover of Plant Physiology, Volume 170, Issue 1, January 2016]
Cellulose is the major structural component of plant cell walls and has great potential as a renewable source of energy. The plant cellulose synthesis complex (CSC), also called a “rosette” because of its hexameric appearance in transmission electron microscope (TEM) images, is a large multi-subunit transmembrane protein complex responsible for synthesis of cellulose chains and their assembly into microfibrils.
Despite the importance of cellulose, fundamental properties of the CSC remain unclear. The number of cellulose synthase (CESA) proteins in the CSC and the number of cellulose chains in a microfibril have been debated for years.
Vandavasi et al. report a solution structure of the catalytic domain of CESA1 from Arabidopsis thaliana determined by small-angle scattering that provides experimental evidence for the self-assembly of CESA into a stable trimer. This study strongly supports the “hexamer of trimers” model for the rosette CSC that synthesizes an 18-chain cellulose microfibril as its primary product.
Funding Acknowledgements
Molecular biology and structural characterization: H.O., V.G.V., Q.Z., W.T.H., L.P., U.K., J.C.S., P.L., and L.C., supported by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC, for the U.S. Department of Energy (DOE) under Contract Number DE-AC05-00OR22725. Small-angle scattering (SAS) and computational analysis, performed by H.O., V.G.V., L.P., B.T.N., and C.H.H., supported by Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center (EFRC) funded by the Office of Basic Energy Sciences (OBES). DOE Office of Science. J.M. and D.K.P. support: High Performance Computing Grant from Oak Ridge Associated Universities (ORAU). Bio-SANS is operated by the Center for Structural Molecular Biology at ORNL, supported by Office of Biological and Environmental Research (OBER), DOE Office of Science, Project ERKP291. EQ-SANS at Spallation Neutron Source (SNS) and High Flux Isotope Reactor (HFIR) sponsored by OBES Scientific User Facilities Division, DOE Office of Science, at ORNL. Sai Venkatesh Pingali: assistance with the operation of Bio-SANS and data reduction. Paul Abraham and the BioEnergy Science Center (BESC) proteomics facilities: validation of purified proteins. BESC supported by DOE OBER. Mass spectrometry (MS) analysis carried out by DOE OBER supported Bioenergy Research Center proteomics pipeline. Use of the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory (BNL), supported by OBES, DOE Office of Science, under Contract Number DE-AC02-98CH10886.
Related Links
References
Vandavasi, V.G., D.K. Putnam, Q. Zhang, L. Petridis, W.T. Heller, B.T. Nixon, C.H. Haigler, U. Kalluri, L. Coates, and P. Langan. 2016. “A structural study of CESA1 catalytic domain of Arabidopsis cellulose synthesis complex: evidence for CESA trimers.” Plant physiology 170(1):123-35. [DOI: 10.1104/pp.15.01356]
