Computational Tool Optimizes SANS Experiments
03/27/2023
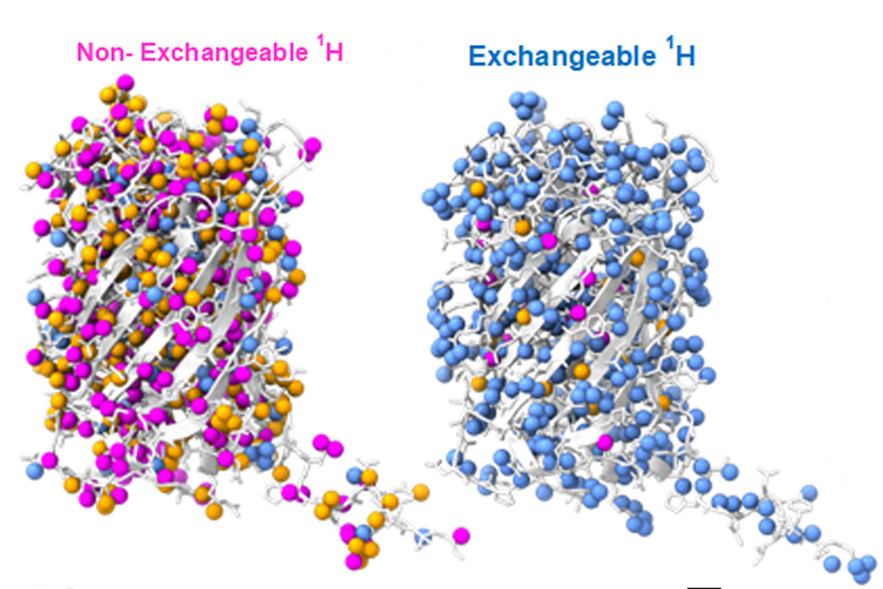
Identifying Deuteration Sites in Biomacromolecules. Both images represent the green fluorescent protein molecule. Non-exchangeable hydrogen sites (pink) and exchangeable hydrogen sites (blue) can be leveraged to alter hydrogen-deuterium ratios that facilitate small-angle neutron scattering (SANS) investigations into biomacromolecular structure.
[Reproduced with permission of the International Union of Crystallography from Hicks et al. (2023). Acta Cryst. D79, 420-434, https://doi.org/10.1107/S2059798323002899.]
The Science
Structural characterization of individual proteins, lipids, and nucleic acids within biomacromolecular complexes can be achieved using small-angle neutron scattering (SANS) and isotopic labeling. To optimize design of SANS experiments, this study developed a contrast matching calculation workflow, called Scattering Contrast Match Points with Explicit-atom Deuteration (SCOMAP-XD), using atomic simulations with explicit-atom deuteration.
Contrast matching is central to SANS because it enables different components of a biological system to be investigated individually. Biological macromolecules are typically dissolved in water containing both hydrogen and deuterium (i.e., heavy water; deuterated water; D2O), which produce different scattering signals. By manipulating the ratios of hydrogen to deuterium in the solution until it progressively reaches match points for different biomolecules, or by manipulating ratios in a specific molecule itself, the system’s different macromolecular components can be observed one at a time.
SCOMAP-XD is a computational tool that explicitly deuterates biomolecular three-dimensional structures to determine the overall contrast match point and the Q-dependent contrast-match points of biomacromolecules. In addition, SCOMAP-XD combines information from empirical models to understand deuterium incorporation at exchangeable and nonexchangeable hydrogen sites in molecules, due to hydrogen-deuterium exchange and the deuterium oxide concentrations in the cell culture media used for biomacromolecule production.
The Impact
SCOMAP-XD calculates the contrast-match points and Q-dependent contrast for experimental optimization of biomacromolecular neutron scattering experiments. Previous methods have only used random assignment or bulk solute contrast effects for determining contrast match points. This approach significantly advances understanding of SANS contrast and enables further investigation into Q-dependent contrast effects. The approach is readily applicable to a large variety of systems, including polymer and carbohydrate systems like cellulose.
The Summary
The SCOMAP-XD method incorporates empirical models for determining the incorporation of deuterium at non-exchangeable hydrogens from the deuterium oxide (D2O) concentration in the culture medium using mass spectrometry and for determining the probability for hydrogen-deuterium exchange at exchangeable hydrogens from nuclear magnetic resonance hydrogen-deuterium exchange data. Using a 3D structure, hydrogen sites for deuterium incorporation can be selected and scattering profiles can be calculated using the explicit solvent calculator, SASSENA. From the calculated structures, the contrast match points can be calculated within 2.5% accuracy. In addition, scattering-vector-dependent contrast effects were observed at high Q in several systems, which are indicative of specific contributions due to deuterium distribution in the sample.
Funding
This work is supported by project ERKPA14 funded by the Biological and Environmental Research program in the U. S. Department of Energy (DOE) Office of Science. Neutron scattering experiments on Bio-SANS were supported by the Center for Structural Molecular Biology, funded by DOE BER project ERKP291. The research used resources at the High Flux Isotope Reactor and Spallation Neutron Source, a DOE Office of Science User Facility operated by Oak Ridge National Laboratory. The research also used resources of the Compute and Data Environment for Science (CADES) at Oak Ridge National Laboratory, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC05-00OR22725.
Related Links
References
Hicks, A., et al. 2023. “SCOMAP-XD: Atomistic Deuterium Contrast Matching for Small-Angle Neutron Scattering in Biology,” Acta Crystallographica Section D 79, 420–34 DOI:10.1107/S2059798323002899
