Controlled Self-Assembly of a Designed Nucleoprotein
11/15/2018
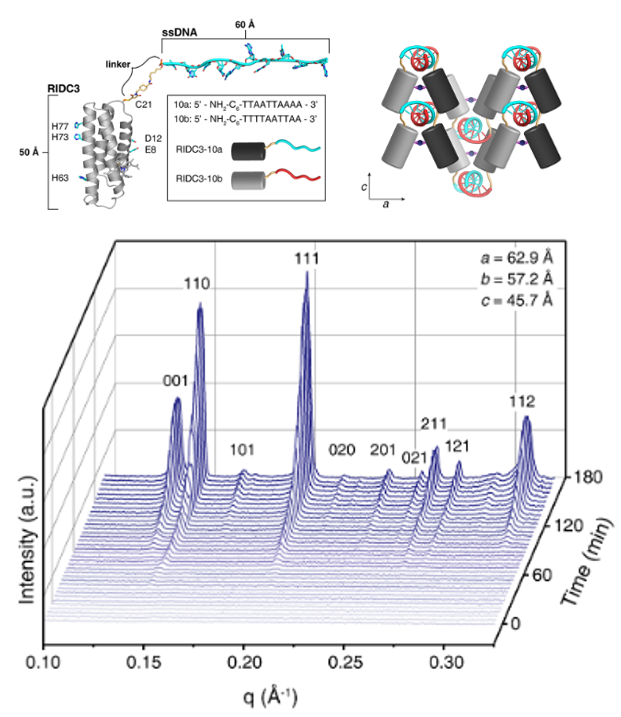
(Top left) Schematic of the protein−nucleic acid hybrid structures RIDC3-DNA. (Top right) Cartoon representation of the RIDC3-DNA three-dimensional (3D) stacking of the 2D crystalline layers. (Bottom) Time-resolved small-angle X-ray scattering (SAXS) profiles of the self-assembly process of RIDC3-10a/b. [Reprinted with permission from Subramanian, R. H., et al. 2018. “Self-Assembly of a Designed Nucleoprotein Architecture through Multimodal Interactions,” ACS Central Science 4, 1578–86. Further permissions related to this material should be directed to the American Chemical Society.]
The Summary
Nature employs a limited set of basic building blocks such as nucleic acids and proteins to create strikingly diverse materials and machines that perform their function with very high fidelity. Inspired by this, synthetic biology and bio-nanotechnology research aims to develop self-assembled structures and materials from the same biological building blocks with functions compatible with or complementary to natural ones that ultimately can surpass those produced by evolution.
In this study, the authors developed a synthetic nucleoprotein assembly that combines a four-helix bundle (RIDC3) with a 10 base pair single-stranded DNA sequence (RIDC3-10a) and its complementary sequence (RIDC3-10b). The construct combines three prominent classes of intermolecular interactions (Watson-Crick base pairing, DNA-protein interactions, and protein-metal coordination with Zn2+) to self-assemble with high structural order and specificity in a manner reminiscent of natural nucleoproteins like the ribosome.
The different structures obtained under a variety of conditions were characterized and structurally identified using equilibrium and in situ time-resolved small-angle X-ray scattering (SAXS) measurements at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 4-2, electron microscopy, and molecular dynamics. The analysis showed that the RIDC3-DNA crystals were discrete two-dimensional (2D) molecular layers that stacked up in 3D (see figure).
While the modular nature of such multicomponent systems should offer distinct advantages in the construction of structurally tunable materials, the intricate architecture of the RIDC3-DNA assembly also highlights the opportunities and challenges inherent in designing artificial nucleoprotein complexes that arise from the distinct structural and chemical properties of proteins and nucleic acids.
Related Links
References
Subramanian, R. H., et al. 2018. “Self-Assembly of a Designed Nucleoprotein Architecture Through Multimodal Interactions,” ACS Central Science 4, 1578−1586. [DOI:10.1021/acscentsci.8b00745.]
