Could This Enzyme Help Turn Biofuel Waste into Something Useful?
04/18/2017
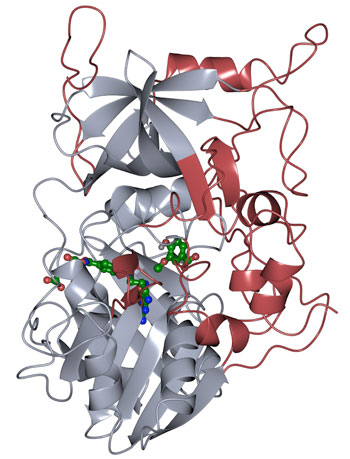
The protein structure of LigM was determined using X-ray crystallography, revealing novel structural elements that are unique to LigM (red) in addition to a conserved tetrahydrofolate-binding domain (gray) that is found throughout life. LigM binds to it’s substrates (green) using internal binding cavities. [Credit: Amanda Kohler/JBEI.]
Joint BioEnergy Institute study targets LigM for its role in breaking down aromatic pollutants
A protein used by common soil bacteria is providing new clues in the effort to convert aryl compounds, a common waste product from industrial and agricultural practices, into something of value. The protein structure of the enzyme LigM was determined using X-ray crystallography, revealing novel structural elements (red in figure) and a conserved tetrahydrofolate-binding domain (gray), with LigM binding to its substrates (green) using internal binding cavities.
Researchers characterized aryl O-demethylation by LigM in Sphingomonas paucimobilis, a soil bacterium that metabolizes aryl compounds derived from lignin—the stiff, organic material that gives plants their structure. In biofuel production, aryl compounds are a byproduct of the breakdown of lignin, some pathways of which involve demethylation, an often critical precursor to additional modification steps of lignin-derived aryl compounds. The simple, single-enzyme system of LigM, as well as its functionality over a broad temperature range, makes it an attractive demethylase for use in aromatic conversion.
Other findings included: half the LigM enzyme was homologous to known structures with a tetrahydrofolate-binding domain that is found in both simple and complex organisms; the other half of LigM’s structure is completely unique, providing a starting point for determining where its aryl substrate-binding site is located; and LigM is a tyrosine-dependent demethylase. This research provides groundwork needed to aid in developing an enzyme-based system for converting aromatic waste into useful products.
Funding Acknowledgements
Crystallographic work: Berkeley Center for Structural Biology (BCSB) Advanced Light Source (ALS) beam line 8.2.2. BCSB ALS staff: technical support; J. H. Pereira: assistance in early stages of crystallographic work. BCSB support in part: National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS) and Howard Hughes Medical Institute (HHMI). ALS support: Office of Basic Energy Sciences (OBES), Director, U.S. Department of Energy (DOE) Office of Science, under Contract DE-AC02-05CH11231. Work conducted by Joint BioEnergy Institute (JBEI) and supported by Office of Biological and Environmental Research (OBER), DOE Office of Science, under Contract DE-AC02-05CH11231.
Instruments and Facilities
Beamline 8.2.2 and X-ray macromolecular crystallography at the Berkeley Center for Structural Biology, Advanced Light Source, Lawrence Berkeley National Laboratory.
Related Links
- BER Resource: Structurally Integrated Biology for the Life Sciences
- Feature Story: Could this enzyme help turn biofuel waste into something useful?
References
Kohler, A. C., et al. “Structure of Aryl O-Demethylase Offers Molecular Insight into a Catalytic Tyrosine-Dependent Mechanism.” PNAS 114(16), E3205–E3214 (2017). [DOI:10.1073/pnas.1619263114]
