Deciphering the Mechanism of Enzymatic Methane Synthesis
03/24/2021
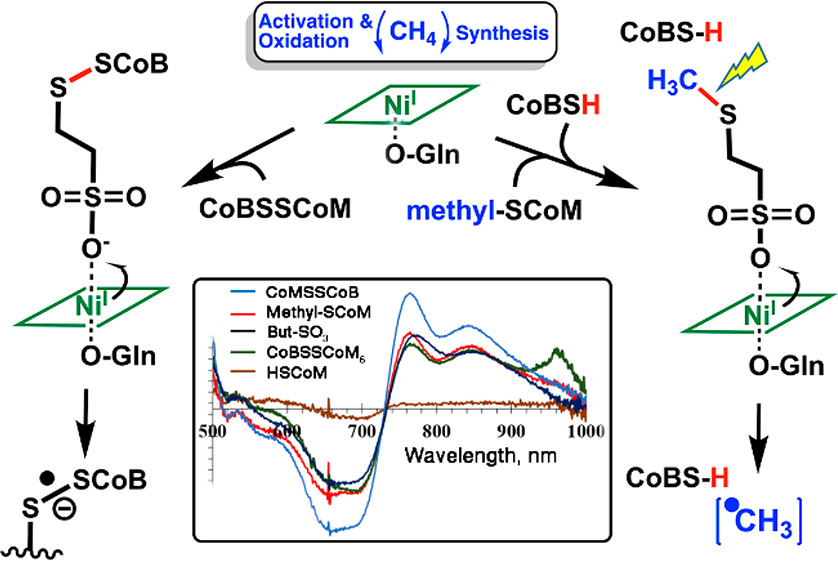
Researchers propose a radical mechanism involving long-range electron transfer based on nickel (Ni) K-edge EXAFS measurements. The measurements reveal a unique 6-coordinate Ni(I)-substrate bound structure previously unseen in biology. [Reprinted with permission from Patwardhan, A., et al. 2021. "Nickel–Sulfonate Mode of Substrate Binding for Forward and Reverse Reactions of Methyl-SCoM Reductase Suggest a Radical Mechanism Involving Long-Range Electron Transfer," Journal of the American Chemical Society 143(14), 5481-5496. Copyright 2021 American Chemical Society.]
The Science
A solution to the substrate-binding conundrum in the mechanism of biological methane synthesis by methyl coenzyme M reductase (MCR).
The Impact
An unprecedented metalloenzyme structure containing nickel (Ni) is believed to initiate methanogenesis via a long-range electron transfer mechanism, expanding the understanding of metal-based catalysis in biology.
Summary
Methane is the simplest organic compound with the highest energy content of any carbon-based fuel. Thus, understanding the biosynthesis of methane is imperative from basic energy, economic and environmental perspectives.
This has led to extensive studies honing in on MCR and its role in catalyzing both the synthesis and anaerobic oxidation of methane. MCR is one of the few Ni-containing proteins in nature, and it is this Ni center that catalyzes the reaction of methyl-coenzyme M (CH3−SCoM) with coenzyme B (HSCoB) to form methane and the heterodisulfide CoMS−SCoB.
Although significant mechanistic studies have been undertaken, none has successfully characterized binding of methyl-SCoM and CoMSSCoB with the active Ni(I) state.
This study describes the coordination chemistry at the active Ni(I) site, elucidating a unique long-range electron transfer process in the MCR reaction mechanism. Clarification of Ni(I)–sulfonate binding requires the field to reassess the previously proposed mechanisms of this important reaction to consider long-range electron-transfer processes.
Research Details
- MCR was generated in its reduced-substrate bound form to differentiate between Ni-S and Ni-O binding modes at the metalloenzyme active site.
- Researchers obtained a solution structure of this protein by extended X-ray absorption fine structure (EXAFS) studies performed on beamline BL7-3 at the Stanford Synchrotron Radiation Lightsource.
- The studies revealed a Ni-O bound structure and provided the critical dataset needed to develop a long-range electron transfer mechanism in methane formation.
- Download technical summary
Funding Acknowledgements
Work in S.W.R.’s group was supported by the US Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES) (Grant DE-FG02–08ER15931). The SSRL Structural Molecular Biology Program was supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences (P30GM133894). Support for S.R. and B.G. was provided by the DOE, Office of Science, Office of Basic Energy Sciences, Division of Chemical Sciences, Geosciences, and Biosciences and was performed in part using the Molecular Sciences Computing Facility (MSCF) in the Environmental Molecular Sciences Laboratory, a DOE User Facility located at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under Contract number DE-AC05-75RL01830.
Related Links
References
Patwardhan, A. et al. 2021 “Nickel–Sulfonate Mode of Substrate Binding for Forward and Reverse Reactions of Methyl-SCoM Reductase Suggest a Radical Mechanism Involving Long-Range Electron Transfer,” J. Am. Chem. Soc. 2021 143 (14), 5481-5496. [DOI: 10.1021/jacs.1c01086]
