Designing a Controllable Self-Assembling Biomaterial
01/06/2021
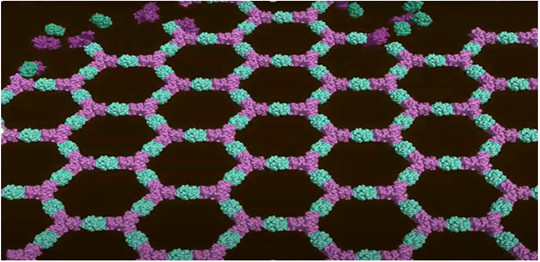
A computationally-designed model of a two-dimensional self-assembling p6m protein lattice comprising a pair of protein building blocks (teal and magenta). Biomaterials composed of two components, rather than a single component, provide greater flexibility in design and assembly and incorporate more complex functionality. [Image courtesy of Lawrence Berkeley National Laboratory.]
The ability to manipulate the components of biomaterials and to control their assembly enables the design of materials with a wide variety of applications.
Most previously described 2D protein materials, such as S-layers and de novo-designed arrays, primarily involve single protein components which spontaneously self-assemble, complicating characterization and repurposing for specific tasks.
To address this problem, scientists have now developed a computational method to generate co-assembling binary layers by designing rigid interfaces between pairs of dihedral protein building blocks. And they used the method to design a hexagonal lattice. Investigation of the kinetics and assembly mechanism in vitro was conducted with high-throughput solution X-ray scattering (small-angle X-ray scattering; SAXS) at the SIBYLS beamline 12.3.1 at the Advanced Light Source.
When combined at nanomolar concentrations, the proteins rapidly assembled into nearly crystalline micrometer-scale arrays almost identical to the computational design model in vitro and in cells without the need for a two-dimensional support.
With this method, protein components can be readily functionalized and their symmetry reconfigured, enabling formation of ligand arrays with distinguishable surfaces. The resulting materials can impose order onto fundamentally disordered substrates such as cell membranes.
Related Links
References
Ben-Sasson AJ, Watson JL, Sheffler W, et al. 2021. “Design of biologically active binary protein 2D materials.” Nature 2021 01; 589(7842):468-473. [DOI: 10.1038/s41586-020-03120-8]
