Designing Cyclic Oligomers: Greater than the Sum of Their Parts
12/05/2016
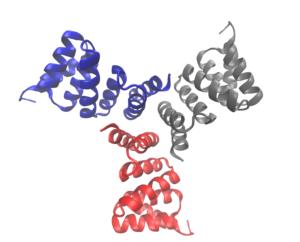
Computational oligomer design: Scientists used small angle solution studies and X-ray scattering to demonstrate that synthetic design matched the computational design. [Reprinted by permission from Springer Nature: Fallas, J. A., et al. 2017. DOI:10.1038/nchem.2673. Copyright 2017.]
The Summary
Cyclic proteins that assemble from multiple identical subunits (homo-oligomers) play key roles in many biological processes, including enzymatic catalysis and function and cell signaling. Researchers in the Molecular Biophysics and Integrated Bioimaging (MBIB) Division worked with University of Washington’s David Baker, who led a team to design in silico and crystallize self-assembling cyclic homo-oligomer proteins.
A strategy was developed that designs interfaces onto idealized proteins aimed to direct their assembly into multimeric complexes. Researchers used structural characterization, both X-ray crystallography and small angle X-ray scattering (SAXS), to show that many of the designs adopted the target oligomerization state and predicted structure. Not only does the work demonstrate that scientists have a basic understanding of what determines oligomerization, but that they could design proteins with tunable shape, size, and symmetry for a variety of biological applications.
Instruments and Facilities
Crystallography Collective program; beamline 5.0.2; small angle X-ray scattering (SAXS) on the SIBYLS BL12.3.1 beamline. Advanced Photon Source synchrotron beamline 24-ID-C at Berkeley Center for Structural Biology and Advanced Light Source at Lawrence Berkeley National Laboratory (LBNL).
Funding
Support: Howard Hughes Medical Institute (HHMI), Air Force Office for Scientific Research (AFOSR FA950-12-10112), National Science Foundation (NSF MCB-1445201 and CHE-1332907), Bill and Melinda Gates Foundation (OPP1120319), and U.S. Department of Defense (DoD) Defense Threat Reduction Agency (HDTRA1-11-C-0026 AM06). R. Koga and L. Carter: assistance with size exclusion chromatography with multi-angle light scattering SEC-MALS. M. Collazo and M. Sawaya support: U.S. Department of Energy (DOE; Grant DE-FC02-02ER63421). M. Capel, K. Rajashankar, N. Sukumar, J. Schuermann, I. Kourinov, and F. Murphy at Northeastern Collaborative Access Team support: grants from National Center for Research Resources (5P41RR015301-10) and National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS; P41GM103403-10). Use of Advanced Photon Source (APS), Argonne National Laboratory (ANL), support: Office of Biological and Environmental Research (OBER), DOE Office of Science, under Contract DE-AC02-06CH11357. X-ray crystallography and SAXS data collected at the Advanced Light Source (ALS, LBNL, Berkeley, CA, DOE Office of Science contract no. DE-AC02-05CH11231); SAXS data collected through the SIBYLS mail-in SAXS program under aforementioned contract no. and funded by DOE OBER Integrated Diffraction Analysis Technologies (IDAT), NIH Minocycline to Improve Neurologic Outcome in Stroke (MINOS; RO1GM105404), and ALS (K. Burnett and G. Hura). Berkeley Center for Structural Biology (BCSB) support in part: NIH NIGMS and HHMI. ALS support: Office of Basic Energy Sciences (OBES), Director, DOE Office of Science, under contract no. DE-AC02-05CH11231.
Related Links
- BER Resource: Structurally Integrated Biology for the Life Sciences
- News: Designing Cyclic Oligomers — Greater Than the Sum of Their Parts
References
Fallas, J. A., et al. “Computational Design of Self-Assembling Cyclic Protein Homo-Oligomers.” Nat. Chem. 9, 353–360 (2017). [DOI:10.1038/nchem.2673]
