Directed Evolution Mimics Allosteric Activation of Enzymes
08/30/2018
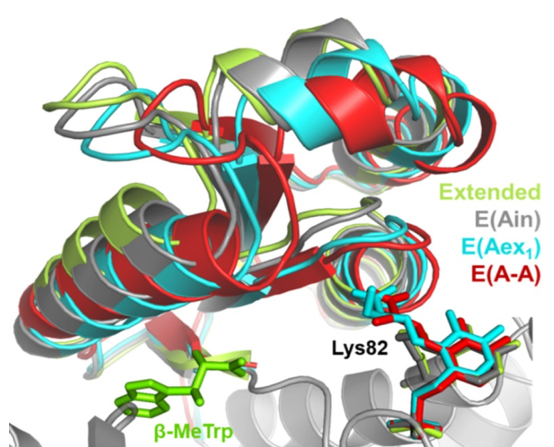
Overlay of tryptophan synthase structures in different states, showing the correlation between the conformational state of the flexible domain and the chemical state of the active site. The extended-open state (green) was observed when a single molecule of β-methyl tryptophan serendipitously bound in a cleft connecting the active site to solvent. The open state in the absence of a ligand bound is shown in gray. A partially closed conformation is observed for externally serine (blue). The formation of the intermediate is coupled to the formation of a fully closed conformational state (shown in red). [Reprinted with permission from Buller, A. R., et al. 2018. “Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble,” Journal of the American Chemical Society 140, 7256–66. Copyright 2018 American Chemical Society.]
The Summary
Biocatalysis is emerging as a powerful methodology for green, sustainable chemical synthesis. While active-site mutagenesis of nonessential residues is a straightforward way to alter substrate specificity and improve function, residues throughout a protein’s structure can also influence catalytic activity. These remote mutations are reminiscent of allosteric effects, where the binding of a ligand or partner protein (an effector) distant from the active site alters enzyme function. Many, if not most, enzymes are allosterically regulated and are an important source of catalytic diversity. Unfortunately, the removal of the native effector often results in loss of activity and can be a major barrier to the successful implementation of enzymes as biocatalysts.
This scenario is exemplified by the enzyme tryptophan synthase, which is a model system for understanding allosteric function as well as a highly desirable biocatalyst for the synthesis of noncanonical amino acids. Nobel Laureate Frances Arnold (California Institute of Technology) and colleagues solved the high-resolution crystal structures of the (1) wild-type enzyme, (2) an intermediate in the lineage, and (3) the final variant, all using the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2.
The structures revealed that the activating mutations have only minor structural effects on their immediate environment; however, they stabilize the large-scale motion of a subdomain to favor an otherwise transiently populated, closed conformational state (see figure). This increase in stability enabled the first structural description of tryptophan covalently bound in catalytically active tryptophan synthase, confirming key features of catalysis.
These data combine to show that sophisticated models are not a prerequisite to mimicking the activation via directed evolution, opening the way to engineering stand-alone versions of diverse allosteric enzymes.
Related Links
References
Buller, A. R., et al. 2018. “Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble,” Journal of the American Chemical Society 140(23), 7256–66. [DOI:10.1021/jacs.8b03490]
