Dynamic Regulation of Histone Chaperone Nucleoplasmin
05/03/2018
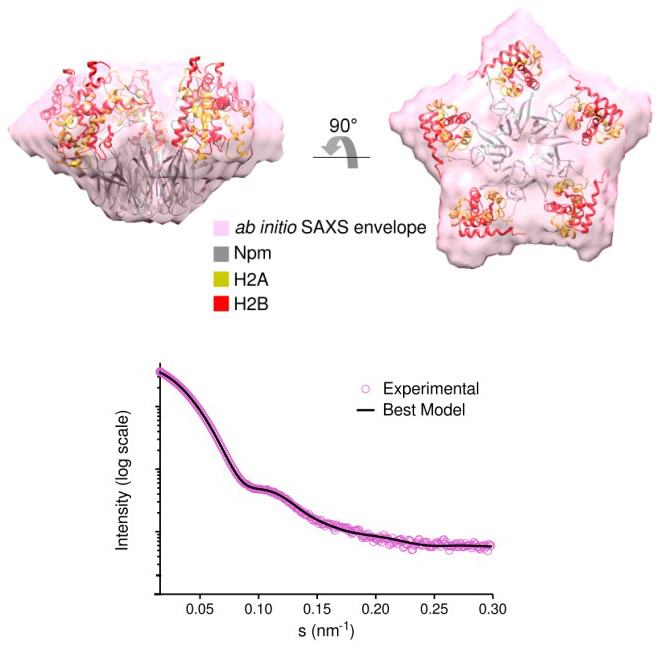
Small-angle X-ray scattering (SAXS) analysis of nucleoplasmin (Npm) Core+A2 truncation (1-145) bound to five H2A/H2B dimers. (Top) SAXS envelope of the pentameric complex (pink) with the best nuclear magnetic resonance (NMR)–restrained SAXS hybrid model inside. (Bottom) SAXS curve of the complex (purple dots). Simulated SAXS curve (black line) from the best-scoring structural model. [From Warren, C., et al. (2017). DOI:10.1038/s41467-017-02308-3. Reused under a Creative Commons license (CC BY 4.0, https://creativecommons.org/licenses/by/4.0/?).]
Histones are eukaryotic cell nuclei proteins that package and order DNA into structural units called nucleosomes. Chromatin is the complex of DNA and proteins comprising the genome’s physiological form. As chromatin’s chief protein components, histones act as spools around which DNA winds, playing a role in gene regulation. A chaperone protein assists in the folding and unfolding of macromolecules, such as in the assembly of nucleosomes from folded histones and DNA. Nucleoplasmin (Npm) is a highly conserved embryonic histone chaperone, responsible for the maternal storage and zygotic release of histones H2A and H2B. Npm contains a pentameric N-terminal Core domain and an intrinsically disordered C-terminal Tail domain. Although intrinsically disordered regions are common among histone chaperones, their roles in histone binding and chaperoning have remained unclear.
This study, using the Xenopus laevis Npm Tail domain, unveils the architecture of the Npm histone complex and a mechanism of histone chaperone regulation. It demonstrates that intramolecular regulation of the histone chaperone Npm controls histone binding and release—a key process in the earliest stages of embryonic development. Structural analyses enabled model constructions of both the Npm Tail domain and the pentameric complex, revealing that the Tail domain controls the binding of histones through specific, electrostatic interactions. Functional analyses demonstrated that these competitive interactions negatively regulate Npm histone chaperone activity in vitro.
Data from these studies establish a potentially generalizable mechanism of histone chaperone regulation via dynamic and specific intramolecular shielding of histone interaction sites.
Instruments and Facilities
Bruker 600 nuclear magnetic resonance (NMR) and Inova 600 NMR instruments in the Albert Einstein College of Medicine (AECM) Einstein Structural NMR Resource; and bio–small-angle X-ray scattering (bio-SAXS) beamline 4-2, SLAC National Accelerator Laboratory’s Stanford Synchrotron Radiation Lightsource (SSRL).
Funding Acknowledgements
Supported by The American Cancer Society (ACS)?Robbie Sue Mudd Kidney Cancer Research Scholar Grant (124891-RSG-13-396-01-DMC) and National Institutes of Health (NIH) grant R01GM108646 (both to D.S.) and training grants T32GM007491 and F31GM116536 (to C.W). J.M.K. supported by Einstein Medical Scientist Training Program Grant (T32 GM007288). Bruker 600 nuclear magnetic resonance (NMR) instrument purchased using funds from NIH award 1S10OD016305 and supported by Albert Einstein College of Medicine (AECM). Inova 600 NMR instrument in the Einstein Structural NMR Resource purchased using funds from NIH award 1S10RR017998 and National Science Foundation (NSF) award DBI0331934 and supported by the AECM. Use of Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory (SLAC), supported by the Office of Basic Energy Sciences (OBES), U.S. Department of Energy (DOE) Office of Science, under Contract No. DE-AC02-76SF00515. SSRL Structural Molecular Biology Program supported by the Office of Biological and Environmental Research (OBER), DOE Office of Science, and by National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS; including P41GM103393).
Related Links
References
Warren, C., et al. “Dynamic intramolecular regulation of the histone chaperone nucleoplasmin controls histone binding and release.” Nat. Commun. 8, 2215 (2017). DOI:10.1038/s41467-017-02308-3.
