Engineering Better Plant Cultivars for Iron Uptake by Modifying the Iron Deficiency Response in Arabidopsis thaliana
06/16/2017
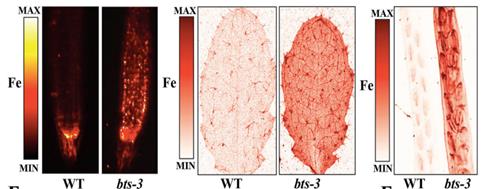
(Left) Representative synchrotron X-ray fluorescent (SXRF) scans of root tips of plants grown for 7 days on B5 medium and then transferred to iron (Fe) medium for 3 days. (Middle) SXRF scan of leaves showing Fe (red). (Right) SXRF scan showing Fe (red), zinc (green), manganese (blue) localization in developed, green siliques. [Reproduced from Hindt, M. N., et al. (2017) with permission of The Royal Society of Chemistry. DOI: 10.1039/C7MT00152E.]
Many populations in developing countries rely on plants for dietary iron (Fe) which is essential for plant growth, crop yields, and human health. However, due to its low or limited solubility, Fe is sparingly available in neutral or basic soils and thus not readily accessible in the rhizosphere. This low solubility leads to restricted Fe content in many plants and is a major factor contributing to the widespread prevalence of Fe deficiency anemia in people with plant-based diets.
Increasing plant Fe acquisition and storage may have profound impacts on plant and human nutrition and can be achieved by manipulating genes and related mechanisms governing Fe homeostasis in plants. However, understanding the balance between positive and negative regulation of the Fe deficiency response is essential for efforts to engineer plants having a sufficient but not toxic level of Fe. Although plants often are challenged with Fe deficiency, no environment remains constant, making Fe availability in the rhizosphere dependent on many factors. When sufficient Fe is available, plants must effectively suppress their Fe-deficiency response to avoid excessive uptake.
A research team has identified a novel Fe-binding domain allele called BTS in a mutagenesis screen for altered Fe accumulation (bts-3). Data showed that bts-3 is more tolerant than wild type to Fe-deficient conditions and that bts-3 is sensitive to Fe-sufficient conditions and accumulates excessive Fe. A triple mutant with loss of both BTS paralogs and a partial loss of BTS expression exhibits even greater tolerance to Fe-deficient conditions and increased Fe accumulation without any resulting Fe toxicity effects, with the mutations also changing their uptake of important minerals such as zinc (Zn) and manganese (Mn). Genetic knockdowns and modifications of the proteins have been implicated in regulating plant uptake of Fe.
This work will lead to greater understanding of plant Fe homeostasis to inform efforts for improved crops. Identifying natural variants of these genes in crop species may lead to traditional breeding efforts to generate higher-Fe cultivars.
Instruments and Facilities
PerkinElmer LAS Ltd, Seer Green, United Kingdom, and Elemental Scientific Inc., Omaha, Neb.; synchrotron X-ray fluorescence (SXRF) at National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory; Stanford Synchrotron Lightsource (SSLS) at SLAC National Accelerator Laboratory (SLAC); real-time quantitative PCR (Step One Plus Real Time PCR System using Applied Biosystems Version 2.2.3; Advanced Photon Source (APS) at Argonne National Laboratory (ANL); and Australian Synchrotron, Victoria. Two-dimensional SXRF analysis was performed at various X-ray microprobe beamlines: microarray analysis and X-ray fluorescence imaging (XFI) on SSLS beamline 2-3 at SLAC; X26A and X27A of NSLS; XFM beamline of the Australian Synchrotron; APS beamline 2-ID-D. Microarray analysis performed at Geisel School of Medicine in the Genomics Shared Resource at Dartmouth College. Elemental concentration analysis (inductively coupled plasma-mass spectrometry (ICP-MS, PerkinElmer NexION 300D equipped with Elemental Scientific Inc. autosampler and Apex HF sample introduction system at PerkinElmer LAS Ltd, Seer Green, U.K., and Elemental Scientific Inc., Omaha, Neb., respectively, in the standard mode.
Funding Acknowledgements
Funding: National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH), U.S. Department of Health and Human Services (HHS) (P42 ES007373); National Cancer Institute (NCI), NIH, HHS (P30 CA023108); National Institute of General Medical Sciences (NIGMS), NIH, HHS (R01 GM078536, P41 GM103393). Microarray analysis: carried out at Geisel School of Medicine in the Genomics Shared Resource, established by equipment grants from NIH and National Science Foundation (NSF); supported in part by a Cancer Center Core Grant (P30CA023108) from NIH NCI. X26Asupport: U.S. Department of Energy (DOE) Geosciences (DE-FG02-92ER14244 to The University of Chicago’s Center for Advanced Radiation Sources (CARS). NSLS support: DOE under Contract No. DE-AC02-98CH10886. X27A support in part: DOE Geosciences (DE-FG02-92ER14244 to CARS and Brookhaven National Laboratory’s (BNL) Department of Environmental Sciences. NSLS support: Office of Basic Energy Sciences (OBES), DOE Office of Science under Contract No. DE-AC02-98CH10886. Sam Webb and Benjamin Kocar: help at Beamline 2–3, Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory (SLAC). Use of SSRL, SLAC support: OBES, DOE Office of Science, under Contract No. DE-AC02-76SF00515. SSRL Structural Molecular Biology Program support: DOE Office of Biological and Environmental Research (OBER) and NIH National Institute of General Medical Sciences (NIGMS; including P41GM103393). Suna Kim and Louisa Howard: aid in in preparation of leaf sections for Figure 4E. Tony Lanzirotti: aid at Advanced Photon Source (APS), an Office of Science User Facility operated for DOE Office of Science by Argonne National Laboratory (ANL), supported by DOE under Contract No. DE-AC02-06CH11357. Martin DeJonge and Daryl Howard: aid at x-ray fluorescence microscopy (XFM) beamline, Australian Synchrotron, Victoria, Australia. Work supported by grants to M.L.G. from NSF (DBI 0701119, IOS-0919941), NIH (R01GM078536), DOE (DE-FG-2-06ER15809), and NIH NIEHS (P42 ES007373), and NSF Plant Genome grant (DBI 0701119) to D.E.S. and M.L.G. M.N.H. support: NSF Graduate Research Fellowship, Nell Mondy Fellowship from Sigma Delta Epsilon-Graduate Women in Science, and Dartmouth Graduate Alumni Research Award.
Related Links
References
Hindt, M. N., et al. “BRUTUS and its Paralogs, BTS LIKE1 and BTS LIKE2, Encode Important Negative Regulators of the Iron Deficiency Response in Arabidopsis thaliana.” Metallomics 9(7), 876–890 (2017). [DOI:10.1039/C7MT00152E].
