Enhancing Enzyme Catalysis Through Directed Evolution
11/19/2020
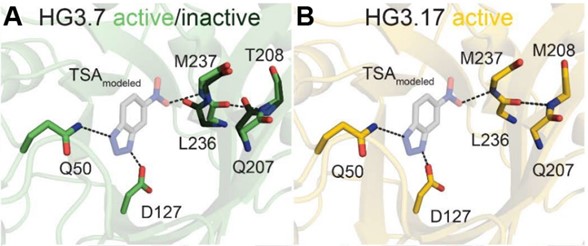
X-ray data reveal extensive structural changes in the artificial Kemp eliminase enzyme (HG3) during directed evolution, enhancing its specific activity. In HG3.7 (A), the active conformation of active site residues (green sticks) makes favorable interactions with a modeled transition state analogue (TSA) (transparent gray) and is stabilized by the conformation of residues Q207-T208. The inactive conformation (black sticks) is binding-incompetent as the carbonyl-group of L236 would clash with the TSA. In contrast, in HG3.17 (B), only the active state of both strands is observed, which enhances the specific activity of the enzyme over 200-fold compared to HG3. [From Otten, R., et al. 2020. “How Directed Evolution Reshapes the Energy Landscape in an Enzyme to Boost Catalysis,” Science 370, 6523, 1442-1446. [DOI:10.1126/science.abd3623]. Reprinted with permission from AAAS.]
Computational enzyme design has aided the development of catalysts for chemical reactions ranging from ester hydrolysis to cycloadditions. Although the starting activities for these enzymes are usually low, they can be increased to levels approaching those of natural enzymes through laboratory evolution. This process mimics the natural selection of enzymes in biology, with the advantage that individual intermediates along the evolutionary pathway can be characterized to deduce how function was enhanced.
For the computationally designed Kemp eliminase HG3 enzyme, researchers wanted to better understand the molecular origins of a rate enhancement achieved by directed evolution. The Kemp elimination, a well-studied model based on a basic xylanase scaffold for proton transfer from carbon, has served as a benchmark for de novo enzyme design. The research team—which included collaborators from the Swiss Federal Institute of Technology, Brandeis University, University of Massachusetts, and Stanford Synchrotron Radiation Lightsource (SSRL)—obtained two evolutionary intermediates from HG3: HG3.7 after seven rounds of mutagenesis and screening and HG3.17 after 17 rounds. The team analyzed these intermediates using nuclear magnetic resonance and x-ray crystallography. The HG3.17 intermediate showed a 200-fold increase in reaction rate compared to HG3. Researchers determined crystal structures of HG3, HG3.7, and HG3.17 using diffraction data collected at SSRL beamlines 14-1 and 7-1 at multiple temperatures and within novel graphene crystal mounts.
The team found that HG3 and HG3.7 both had dual conformers near the active site, one of which can destabilize the transition state. However, in HG3.17, only one confomer existed, which stablizes the transition state. This study shows that explicit screening of productive conformations that confer better enzymatic activity can be used to improve design protocols and guide development of future mutagenesis and screening strategies.
Related Links
References
Otten, R., et al. 2020. “How Directed Evolution Reshapes the Energy Landscape in an Enzyme to Boost Catalysis,” Science 370, 6523, 1442-1446. [DOI:10.1126/science.abd3623]
