Hacking the Bacterial Social Network
08/11/2017
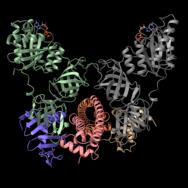
This representation shows the neutralized complex of the CdiA toxin (purple and beige) with the CdiI immunity protein (orange and pink) and the elongation factor (EF)–Tu (grey and green). Scientists have determined the molecular structure of this protein complex—an insight that could lead to new biomedical strategies for overcoming pathogenic bacteria. [Karolina Michalska, Argonne National Laboratory]
Scientists have determined the molecular structures of a highly specialized set of proteins that a strain of Escherichia coli bacteria use to communicate and defend their turf. The research shows that bacteria appear to have a social network that lets them attract and repel one another, potentially leading to new biomedical strategies for overcoming pathogenic bacteria that cause infectious diseases such as pneumonia and food-borne illnesses.
The work builds on the 2005 discovery that bacteria produce toxic proteins which they can transfer to their neighbors through direct contact to either kill or control them. This strategy may help them gain better access to nutrients in densely populated microbial communities through a process called contact-dependent growth inhibition (CDI). Learning how bacteria interact and communicate is helping to resolve the different activities of the toxins, which may affect different bacteria differently. CDI system toxins are found in soil and gut bacteria, as well as in human pathogens like Pseudomonas aeruginosa which is involved in lung disease.
The research team used high-bright X-rays to characterize biological proteins and inspect chemical processes at the nanoscale level, obtaining the molecular structures of proteins that belong to a three-part system of the NC101 strain of E. coli. The system consists of the CDI toxin, its immunity protein, and its elongation factor (EF). The latter, known as EF-Tu, is a protein that plays a key role in protein synthesis. Knowing the protein structures of all three parts helps scientists understand their function.
Discovery of the immunity protein has led scientists to suspect that the system’s purpose includes competition and signaling (intracommunication), as well as killing and controlling other bacteria. Only a few molecules of the toxin reach the neighboring cell, so perhaps the toxin is not meant to kill, but rather to control and communicate. The toxin can act on transfer ribonucleic acid (tRNA) only under highly specific circumstances and it is the first case seen where EF is needed for the toxin to function.
Instruments and Facilities
Advanced Photon Source 19-ID beamline and Advanced Protein Characterization Facility at Argonne National Laboratory.
Funding Acknowledgements
National Institutes of Health (NIH; GM094585, GM115586 to A.J.; GM117373 to C.W.G., D.A.L., C.S.H.); Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE) Office of Science [DE-AC02-06CH11357 to A.J.]. Funding for open access charge: NIH grant [GM117373].
Related Links
References
Michalska, K., et al. “Structure of a Novel Antibacterial Toxin That Exploits Elongation Factor Tu to Cleave Specific Transfer RNAs.” Nucleic Acids Res. 45(17), 10306–10320 (2017). [DOI:10.1093/nar/gkx700].
