Hollow Pyramid Unlocks Principles of Protein Architecture
12/14/2016
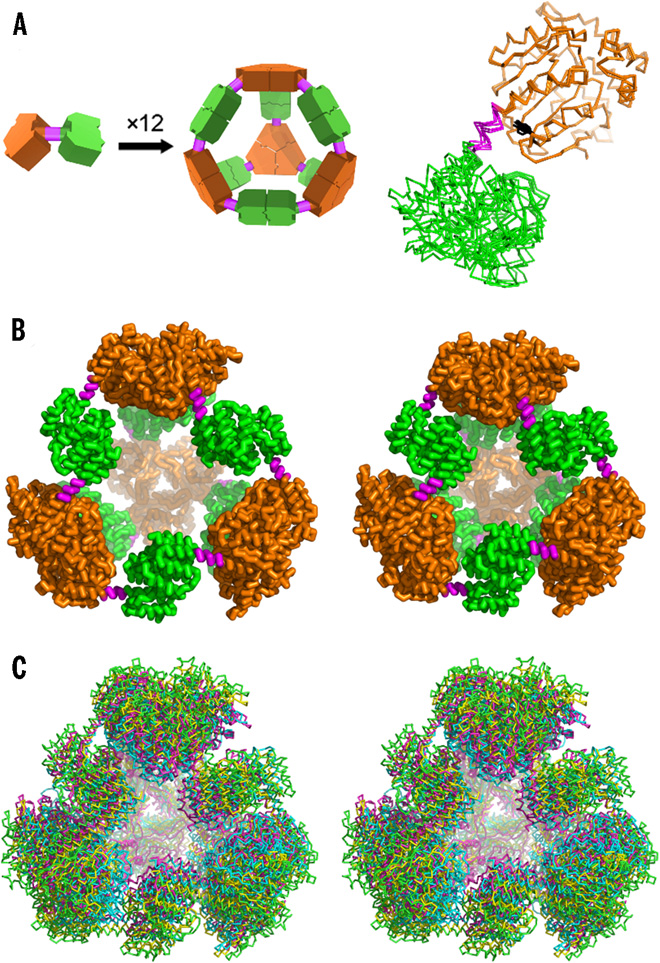
Designed and observed cage symmetry. (A) Left: Schematic diagram of the symmetry principles used to design the 12-subunit tetrahedral cage by fusing two oligomeric domains (green and orange) by a semirigid linker (magenta). Right: Single point mutation distinguishing our PCtrip construct from PCquad replaces tyrosine (black sticks) with alanine in the trimeric domain that makes contact with the linker. (B) Side-by-side view of the theoretically designed (perfectly symmetric) model of the protein cage (left) and the most symmetric crystal structure obtained in this work for the PCquad variant (right). (C) Walleyed stereo view of three crystal structures of PCquad (yellow, magenta, and blue) overlaid on the ideal model (green ribbon), showing the agreement of the observed structures and the design. [[From Lai, Y.-T., et al., “Designing and Defining Dynamic Protein Cage Nanoassemblies in Solution.” Sci. Adv. 2(12), e1501855 (2016). DOI:10.1126/sciadv.1501855. Reused under a Creative Commons license (CC BY NC-4.0, https://creativecommons.org/licenses/by-nc/4.0/.]
Large biological macromolecules (up to 10,000 atoms) can catalyze chemical reactions in ways that are difficult to replicate inorganically. Their size allows for the complexity needed to perform such functions, but it also makes them more susceptible to misfolding and aggregating—thus, the importance of understanding the fundamental architectural principles that cause large proteins to favor specific conformations. At the nanoscale, where organic chemical groups interact with solvent water molecules, these principles are very different from the ones used to build houses and cars. To investigate these principles, a group of researchers designed a protein that would self-assemble into a hollow pyramid (or tetrahedron). Upon crystallizing the macromolecule, the group found that they had indeed been successful in creating the assembly, but it was unexpectedly warped and collapsed in an asymmetric manner, with some edges bent inward—an asymmetric tetrahedron. To ensure that this was not an artifact of crystallization, they investigated the protein’s behavior in solution using small-angle x-ray scattering (SAXS), which showed that the collapse could be controlled by adjusting the solution’s salt concentration; the structure was disassembled by varying the pH.
The flexibility of this macromolecule suggests that it could be useful for the controlled capture and release of smaller compounds. Overall, the researchers expect that, with the tools and techniques developed here, the combination of SAXS with crystallography or electron microscopy could be increasingly useful in analyzing and optimizing designed protein assemblies and understanding their behavior in solution.
Instruments and Facilities
Small-angle X-ray scattering (SAXS) at Advanced Light Source (ALS) with SIBYLS Beamline 12.3.1 (a joint crystallography and SAXS beamline) at Lawrence Berkeley National Laboratory.
Funding Acknowledgements
Berkeley Center for Structural Biology (BCSB), Advanced Light Source (ALS), Lawrence Berkeley National Laboratory (LBNL), work performed as collaboration between the Joint BioEnergy Institute (JBEI; https://www.jbei.org) and Great Lakes Bioenergy Research Center (GLBRC; https://www.glbrc.org). JBEI support: Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE) Office of Science, through contract DE-AC02-05CH11231 between LBNL and DOE. GLBRC support: OBER, DOE Office of Science, through Grant DE-FG02-07ER64495. BCSB support in part: National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS). ALS support: Director, Office of Basic Energy Sciences (OBES), DOE Office of Science, under Contract DE-AC02-05CH11231. Support for part of work: National Science Foundation (NSF) under Cooperative Agreement 1355438.
Related Links
- BER Resource: Structurally Integrated Biology for the Life Sciences
- Feature Story: A Hollow Pyramid Unlocks Principles of Protein Architecture
References
Lai, Y.T., G.L. Hura, K.N. Dyer, H.Y. Tang, J.A. Tainer, and T.O. Yeates. 2016. “Designing and defining dynamic protein cage nanoassemblies in solution.” Sci Adv 2(12) e1501855. [DOI: 10.1126/sciadv.1501855]
