Developing an E.coli Platform for Hydrogenase Production
07/21/2023
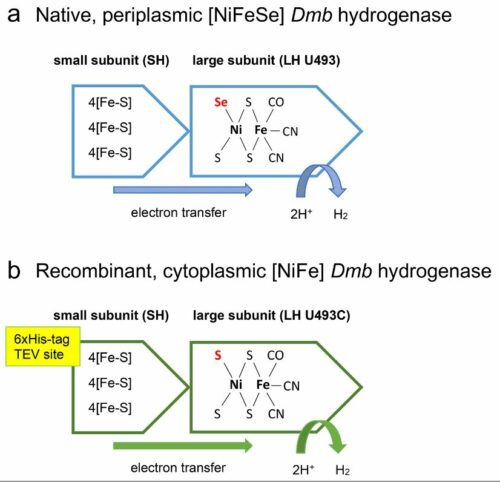
One step closer to industrial biohydrogen production using microbial hydrogenases. The image presents molecular differences between a hydrogenase enzyme produced natively by Desulfomicrobium baculatum (a) and a similar, functional hydrogenase produced by recombinant genes in E. coli (b). Note the substitution of sulfur for selenium (red) in the enzyme’s active site. The yellow box indicates a tag that enables rapid protein purification.
[Reprinted under a Creative Commons license (CC BY) from Witkowska et al. 2023. DOI:10.1186/s12934-023-02127-w]
The Science
Hydrogenases are metalloenzymes, found in microbes, which efficiently convert protons and electrons to molecular hydrogen. Industrial production of hydrogen, a potential fossil fuel alternative, requires intensive use of chemicals, toxic metals, and energy. Developing an industrial process that exploits the unique enzymatic activity of microbial hydrogenases could advance the process of biohydrogen evolution and green energy production.
Researchers from the University of Gdansk and Argonne National Laboratory used site-directed mutagenesis of a bacterial hydrogenase operon—a cluster of genes that work in concert to produce the enzyme—and molecular imaging to create a functional, recombinant operon for rapid and robust hydrogenase production in the industrial powerhouse E. coli. Using such strains alleviates the challenging growth constraints of much slower growing and difficult to cultivate native sources of hydrogenase—such as the anaerobic, sulfate-reducing Desulfomicrobium baculatum used in this study.
The resulting recombinant hydrogenase varies from the native enzyme in that its active site is composed of nickel and iron (Ni-Fe), whereas the native enzyme has a Ni-Fe-Selenium (Ni-Fe-Se) core. The Ni-Fe hydrogenase displays only a fraction of the native enzyme’s activity, and its properties still need to be evaluated, but the study demonstrates a promising strategy for cloning, expressing, and purifying catalytically active hydrogenase derived from D. baculatum in E. coli.
The Impact
The genetic constructs created in this study, together with improved growth and purification procedures, provide a promising platform for further studies toward producing fully active and oxygen-tolerant recombinant Ni-Fe-Se hydrogenase resembling the native D. baculatum enzyme.
The Summary
The native Ni-Fe-Se hydrogenase from D. baculatum was targeted in this study for its natural oxygen tolerance and high hydrogen evolution activity. The native hydrogenase operon includes two structural hydrogenase genes, coding for large and small subunits, and an additional gene, encoding a protease essential for proper enzyme maturation. Recombinant expression of these exact genes in E. coli, however, produces inactive enzymes. Therefore, researchers converted the native Ni-Fe-Se hydrogenase to an Ni-Fe type hydrogenase using site-directed mutagenesis.
The four resulting recombinant hydrogenase variants demonstrated limited ability to produce hydrogen both in vitro and in vivo.
The aim of this study was to overcome bottlenecks in gene expression, protein biosynthesis, maturation, and ligand loading for simple, rapid, and cost-effective delivery of recombinant Ni-Fe hydrogenase using commonly available E. coli strains. Although the enzymes produced in this study do not yet rise to the level of the native enzyme, the demonstrated platform for recombinant hydrogenase production shows promise. Improvements could likely be achieved by selective cysteine to selenocysteine substitution within the active site of the Ni-Fe variant.
Funding
This work was supported by Polish National Science Centre Grant for project titled: Photo-bio hydrogen production by [NiFe] hydrogenase-MNPs/SiO2/MaSb hybrids under visible light irradiation (2016/23/D/ST8/02682), National Centre for Research and Development POWR.03.02.00-IP.08-00-DOK/16, Polish Ministry of Science and Higher Education DS 531-T040-D839-22, DS 531-T040-D839-23, DS 531-T020-D845-23 and in part by National Institutes of Health Grant GM115586 and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02–06CH11357 for the Midwest Center for Structural Genomics Community Resource.
Related Links
References
Witkowska, M., et al. 2023. “Promising approaches for the assembly of the catalytically active, recombinant Desulfomicrobium baculatum hydrogenase with substitutions at the active site,” Microbial Cell Factories 22, 134. DOI:10.1186/s12934-023-02127-w
