Modeling the Partitioning of Amphiphilic Molecules and Cosolvents in Biomembranes
10/14/2022
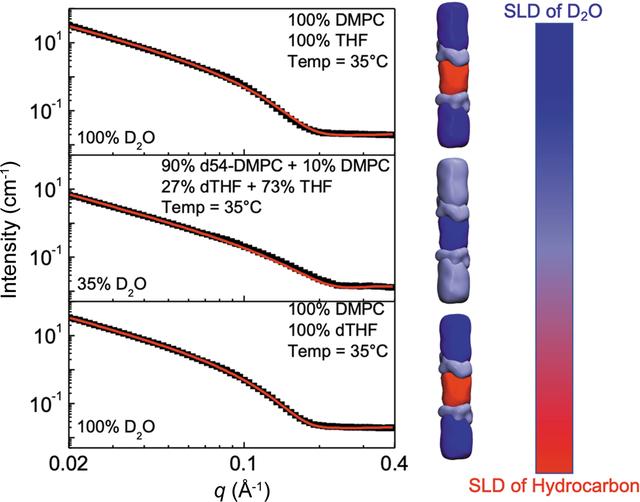
Contrast variation of SANS measurements. Small-angle neutron scattering (SANS) measurements were performed on vesicles comprising the lipid 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) in the presence of the amphiphilic cosolvent tetrahydrofuran (THF). Three separate contrast conditions (left) enabled resolution of different component compositions of the sample, such as the water/heavy water (D2O) ratio in the solvent, the d-THF/THF ratio in the cosolvent, and the deuterated amphiphile lipid/hydrogenated amphiphile lipid ratio. The different contrast conditions are depicted as a color scale to the right of the figure (SLD=scattering length density).[Reproduced with permission of the International Union of Crystallography from Tan et al. 2022. J. Appl. Cryst. 55, 1401-1412.]
The Summary
A primary limitation to fermentation processes is cosolvent and end-product cytotoxicity. Industrial solvents used in lignocellulosic pretreatment and aliphatic fermentation end-products interact with various cellular systems, impacting cell growth and yield in biofuel-producing microbes. The partitioning of small amphiphilic molecules into lipid bilayers from aqueous solution and the corresponding effect on bilayer structure is therefore of fundamental biophysical interest and of significant practical importance.
Small-angle neutron scattering (SANS) is a key method for studying lipid and polymer bilayer structures. In this study, researchers developed a model for analyzing SANS measurements of solvent partitioning in lipid membranes. This is important for quantifying changes in the lipid bilayer structure of cells when exposed to biofuels and other fermentation products such as ethanol, butanol, or acetic acid.
Existing models for interpreting SANS data of lipid membranes have struggled to reliably determine the molecular details of cosolvent partitioning into different regions of the lipid bilayer and changes to the bilayer structure. To address this, researchers developed a model of a bilayer structure with a two-term partition constant accounting for the localization of the cosolvent within the bilayer.
The new model divides the lipid bilayer into three slabs: two for the head groups on either end of the bilayer and a single slab for the central acyl tail section. This model was successfully applied to SANS measurements of lipid vesicles in the presence of tetrahydrofuran (THF), yielding structural information for the bilayer and information about THF partitioning into the bilayer. Model assumptions were evaluated by molecular dynamics simulations. The approach yielded estimates of the partition coefficient for THF in 1,2-dimyristoyl-sn-glycero-3-phosphocholine at 35°C, along with an estimate of the fraction of THF residing in the hydrophobic core of the membrane.
This analysis approach can be applied to many other bilayer/amphiphile interactions. The code needed to implement the model as an algorithm for scattering data has been added to the SASView software suite for small angle scattering data analysis.
Funding
Support was provided by the US Department of Energy (DOE), Office of Science, through the Genomic Science Program, Office of Biological and Environmental Research (contract no. FWP ERKP752). Authors H.L.S. and J.K. are supported through the Scientific User Facilities Division of the DOE Office of Science, sponsored by the Basic Energy Science (BES) Program, DOE Office of Science (contract no. DE-AC05-00OR22725). Neutron scattering research conducted at the Bio-SANS instrument, a DOE Office of Science, Office of Biological and Environmental Research resource (contract no. ERKP291), used resources at the High Flux Isotope Reactor, a DOE Office of Science Scientific User Facility operated by Oak Ridge National Laboratory.
Related Links
References
Tan et al. 2022. Modeling the partitioning of amphiphilic molecules and co-solvents in biomembranes. Journal of Applied Crystallography, 55, 1401-1412. [DOI: 10.1107/S1600576722008998]
