Neutrons Identify Oxygen Activation in LPMO enzyme
12/22/2016
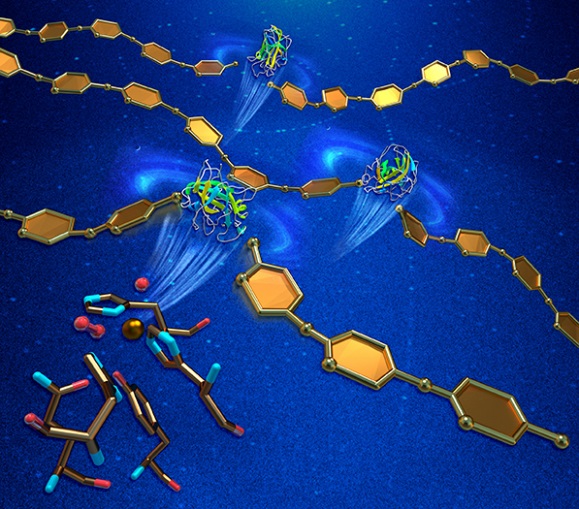
Oxidative cleavage of glycosidic bonds. Neutron and X-ray crystallography reveal structural details of the enzymatic action of lytic polysaccharide monooxygenase (green and yellow ribbon diagrams) as it breaks polysaccharide chains (gold). This activity enhances the enzymatic hydrolysis of recalcitrant carbohydrate biomass, such as cellulose or chitin. [From O’Dell, W. B., P. K. Agarwal, and F. Meilleur. 2017. “Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase,” Angew. Chem. Int. Ed. 56, 767–770. DOI: 10.1002/anie.201610502. © 2017 Wiley‐VCH Verlag GmbH & Co. KGaA, Weinheim.]
Fungal lytic polysaccharide monooxygenases (LPMOs) are known to enhance the efficiency of cellulose-hydrolyzing enzymes through oxidative cleavage of the glycosidic bonds. For this study, PMO-2 from Neurospora crassa was heterologously expressed from Pichia pastoris, purified, and crystallized for high-resolution X-ray crystal structures that revealed “prebound” molecular oxygen in the resting state and a dioxo species in complex with the catalytic copper (Cu2+) ion, which is the first structural description of molecular oxygen (O2) activation by a LPMO. In addition, neutron diffraction studies and density functional theory calculations have identified a role for a conserved histidine in promoting oxygen activation. Extension of these studies to the enzyme-substrate complex could provide a complete picture of the enzymatic mechanism for the potential benefit of applications such as bioethanol production.
Instruments and Facilities
Neutron crystallography. Joint X-ray/neutron refinement at Center for Structural Molecular Biology at Oak Ridge National Laboratory (ORNL). Diffraction data were collected at SER-CAT 22-ID at the Advanced Photon Source at Argonne National Laboratory and at CG-4D IMAGINE (NSF MRI 09229719) at the High Flux Isotope Reactor at ORNL.
Funding Acknowledgements
Protein expression and purification experiments at Center for Structural Molecular Biology (CSMB), a User Facility of the Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE) Office of Science. Diffraction data collected at SER-CAT 22-ID at Argonne National Laboratory’s (ANL) Advanced Photon Source (APS) and at CG-4D IMAGINE (National Science Foundation [NSF] magnetic resonance imaging [MRI] 09229719) at Oak Ridge National Laboratory’s (ORNL) High Flux Isotope Reactor (HFIR), both DOE Office of Biological Energy Sciences (OBES) User Facilities. W.B.O. support: NSF IGERT 1069091. F.M. support: U.S. Department of Agriculture (USDA National Institutes of Food and Agriculture (NIFA) Hatch 211001. P.K.A. support: NIH GM105978.
Related Links
References
O’Dell, W. B., et al. “Oxygen Activation at the Active Site of a Fungal Lytic Polysaccharide Monooxygenase.” Angew. Chem. Int. Ed. 129(3), 785–788 (2017). [DOI:10.1002/anie.201610502].
