Phenoxy Radical-Radical Coupling in Plants
08/14/2020
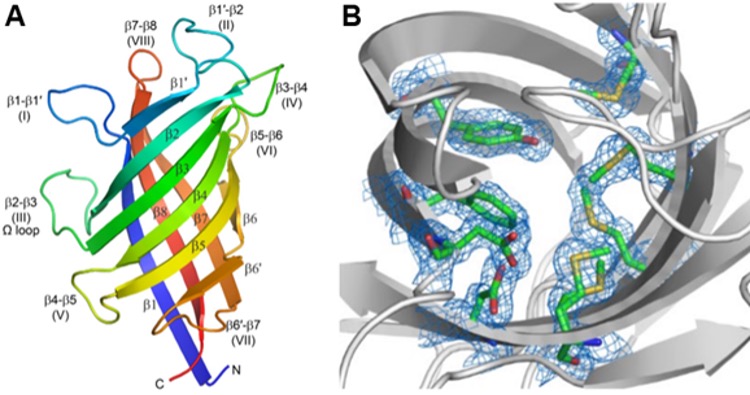
Monomeric Structure of Pterocarpan Synthase from Pisum sativum. (A) The structure, determined using X-ray macromolecular crystallography, is an 8-stranded β-barrel composed of two curved anti-parallel sheets. The barrel is colored from the N-terminus (blue) to the C-terminus (red). The opening to the active site is at the top of the barrel in this orientation, surrounded by flexible loops. (B) The view down into the active site (gray ribbons) shows the residues that make up the active site (green sticks) and their associated electron density as a blue mesh contoured at 1σ. Structural characterizations such as this provide insights into the biochemical mechanisms underlying the formation of pterocarpans and other plant compounds associated with phenoxy radical-radical coupling reactions. [Meng et al. 2020. DOI:10.1074/jbc.RA120.012444]
The Science
The evolution of phenolic radical-radical coupling in plants was pivotal in the transition of plants from early aquatic forms, such as algae and cyanobacteria, to the huge diversity of terrestrial plants today. The reactions form carbon-carbon and carbon-oxygen bonds via radical formation in phenolate compounds, and the molecules ultimately produced are responsible for defense, structural integrity, and reinforcement of plants ranging from grasses and ferns to the largest trees. These reactions generate compounds such as lignans, lignins, pterocarpans, and gossypol, which together account for 30% to 50% of vascular plant mass and represent a massive sink for sequestered carbon.
Research to understand the formation of these compounds has found that the first step in lignan biosynthesis is the stereospecific coupling of two molecules of coniferyl alcohol. Studies revealed two isomers of the lignan product pinoresinol in plants, but how these reactions were catalyzed was not fully understood until researchers from Washington State University identified dirigent proteins (DPs) in the late 1990s. DPs evidently emerged during plants’ transition from water to land, with phylogenetic analyses revealing numerous DP subfamilies throughout the plant kingdom. The biochemical functions of most of these DPs (>95%) still are unknown.
The Impact
To better understand DP biochemical activities and how they give rise to distinct classes of plant phenolic compounds, research teams examined DP structures using the capabilities of the Structural Molecular Biology program at the Stanford Synchrotron Radiation Lightsource (SSRL). Results will help inform predictions and efforts to identify the metabolic pathways and precise biochemical roles that DPs play in the coupling reactions that lead to important plant compounds.
Summary
In 2015, researchers used data collected at SSRL beamline 12-2 to solve the structure of the DP that forms (+)-pinoresinol, a widely distributed plant lignan (Kim et al. 2015). The structure was solved as a dimer of two β-barrel molecules in the asymmetric unit, but there is a very tight trimer generated by a threefold axis. Despite the oligomeric unit most likely being a trimer, each monomer acts independently, and each has an isolated active site at one end of the β-barrel.
Recently, Meng et al. (2020) used data collected at beamline 9-2 to determine the crystal structures of two additional DPs that catalyze the formation of medicarpin, a pterocarpan precursor (see figure). Docking simulations and analysis of the substrate binding sites show how the diastereomeric chiral isoflavonoid precursors might bind and how the radical formed during the subsequent coupling reaction would be stabilized. Moreover, Washington State University, SSRL, Pacific Northwest National Laboratory, and the U.S. Department of Energy’s Joint Genome Institute recently established a joint research program to generate a large number of DPs of varying function. Each DP will be characterized enzymatically and structurally to gain a better understanding of the range and variability of the coupling reactions catalyzed by these important enzymes.
Related Links
References
Kim, K.-W., et al. 2015. “Trimeric Structure of (+)-Pinoresinol-Forming Dirigent Protein at 1.95 Å Resolution with Three Isolated Active Sites,” Journal of Biological Chemistry 290, 1308–18. [DOI:10.1074/jbc.M114.611780]
Meng, Q., et al. 2020. “Pterocarpan Synthase (PTS) Structures Suggest a Common Quinone Methide-Stabilizing Function in Dirigent Proteins and Proteins with Dirigent-Like Domains,” Journal of Biological Chemistry 295, 11584–601. [DOI:10.1074/jbc.RA120.012444]
