Protein Design for Genetically Encodable Nanomachines
04/21/2022
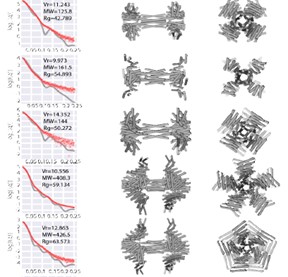
Small-angle X-ray scattering (SAXS) and cryo-EM were used to confirm that constructed axle-rotor protein complexes matched designs. The complexes are intended to eventually become components in genetically encodable nanomachines. SAXS data (left) were collected from nearly 100 partial and full axle-rotor assembly constructs. The SAXS data was compared to SAXS calculated from designs (two perpendicular orientations shown on right). Successful matches were then further validated using cryo-EM. [From Courbet et al. 2022. Computational design of mechanically coupled axle-rotor protein assemblies. Science, 376(6591):383-390. Reprinted with permission from AAAS.]
The Summary
Natural molecular machines contain protein components that undergo motion relative to each other. Designing such mechanically constrained nanoscale protein architectures is an outstanding challenge for computational protein design. Scientists explored the de novo construction of protein machinery from designed axle and rotor components that display internal cyclic or dihedral symmetry.
Small-angle X-ray scattering (SAXS), at the DOE-supported Structurally Integrated Biology for the Life Sciences resource (SIBYLS), was used to screen nearly 100 axle and rotor constructs that would eventually be assembled to form axle-rotor systems (see figure). Once assembled, cryo-EM was used to verify that the constructs matched the design models. This combined approach confirmed that the axle-rotor systems assembled in vitro and in vivo as designed. Achieving construction of mechanical systems with internal degrees of freedom is a step toward the design of genetically encodable nanomachines.
These proof-of-concept axle-rotor assemblies demonstrate that protein nanostructures with internal mechanical constraints can now be systematically designed. Key to this advance is the ability to computationally design protein components with complex complementary shapes, symmetries, and topologies, such as high-aspect-ratio dihedral axle structures (D2 homotetramers to D8 homo-16-mers) with oligomerization states and sizes considerably larger than previously designed.
Efficient techniques like SAXS are necessary to validate computational design. Studying the assembly of shape-complementary homo-oligomeric components into higher-order hetero-oligomeric structures with internal degrees of freedom provides insight into further design of complex protein nanomachines.
Funding
SAXS data were collected at the Advanced Light Source (ALS) SIBYLS 12.3.1 beamline on behalf of U.S. DOE-BER, through the Integrated Diffraction Analysis Technologies (IDAT) program. This research used resources of the ALS, a US DOE Office of Science User Facility under contract DE-AC02-05CH11231. Some of this work was performed at the Pacific Northwest Center for Cryo-EM (PNCC), which was supported by NIH grant U24GM129547 and performed at the PNCC at Oregon Health & Science University and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. A full listing of funding sources can be found in Courbet et al. 2022.
Related Links
References
Courbet et al. 2022. Computational design of mechanically coupled axle-rotor protein assemblies. Science, 376(6591):383-390. [DOI: 10.1126/science.abm1183]
