RuBisCO Evolution Enabled by Structural Plasticity
08/26/2022
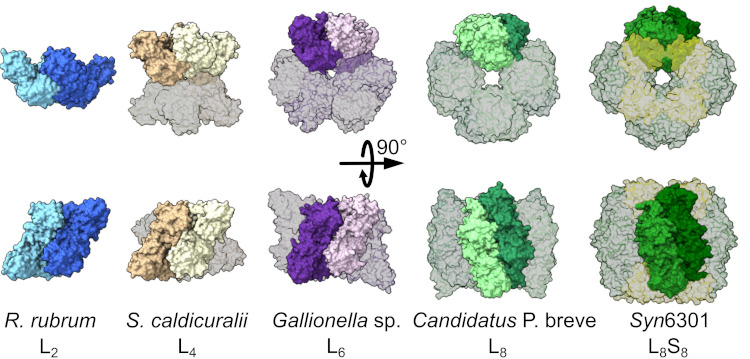
A comparison of RuBisCO assemblies from different microbial species illustrates the enzyme’s wide diversity of oligomeric states. From left to right: form II dimer of Rhodospirillum rubrum, form II tetramer of Sulfurivirga caldicuralii, form II hexamer of Gallionella sp., form I′ octamer of Candidatus Promineofilum breve, and form I hexadecamer of Synechococcus elongatus PCC 6301. [Reprinted under a Creative Commons Attribution License 4.0 (CC BY 4.0) from A.K. Liu et al. 2022. DOI: 10.1126/sciadv.adc9440]
Researchers from Lawrence Berkeley National Laboratory studying the evolution of Ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) reveal that the enzyme’s structural plasticity underlies its rich diversity of molecular assemblies. RuBisCO, the world’s most abundant protein, catalyzes the first step in carbon fixation.
To better understand the phylogenetic distribution of the oligomeric states of form II RuBisCO, found in bacteria and certain photosynthetic microbes, the researchers structurally characterized 28 candidates spread across the phylogeny using traditional macromolecular X-ray crystallography (MX) and small-angle X-ray scattering (SAXS). SAXS offers lower resolution than MX but can take snapshots of proteins in their native form when suspended in solution.
Combining this structural data with respective protein-coding gene sequences enabled the team to also perform ancestral sequence reconstruction. The reconstruction suggested that form II RuBisCO structure has remained plastic throughout its evolutionary history.
To further test the theory that structural plasticity can give rise to new forms, the researchers performed mutational experiments, finding that functional changes in oligomerization could be produced with surprisingly few mutations. In some cases, the new RuBisCO assemblies exhibited improved affinity for their target molecule, carbon dioxide. The results suggest that form I RuBisCO engineering efforts in plants likewise may not need to focus exclusively on the enzyme’s active site to develop more productive and resource-efficient crops.
X-ray scattering experiments were performed using the SIBYLS beamline at the Advanced Light Source and X-ray crystallography was performed at the Berkeley Center for Structural Biology. Research was also supported by the DOE Joint BioEnergy Institute and the Integrated Diffraction Analysis Technologies (IDAT) program.
Related Links
- BER Resource: Structurally Integrated Biology for the Life Sciences
- Feature Story: Protein Structures Aren’t Set in Stone
References
Liu, A.K., et al. 2022. Structural plasticity enables evolution and innovation of RuBisCO assemblies. Science Advances, 8 (34). [DOI: 10.1126/sciadv.adc9440]
