SANS Reveals Role of Disordered Protein Domain
01/24/2023
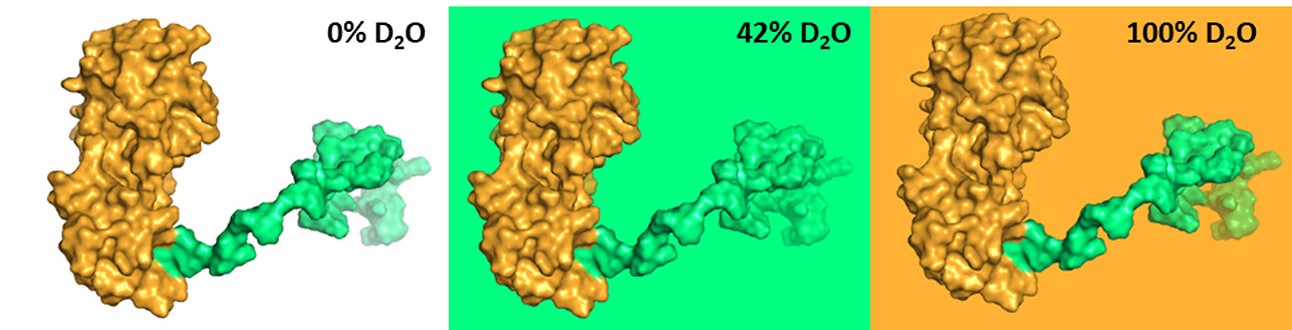
Technique combination zooms in on specific structures. Segmental labeling of c-Src tyrosine kinase domains SH4UD (green) and SH3 and SH2 (gold) enable selective study of individual domains using SANS. In 42% D2O solvent, SH3 and SH2 dominate the SANS signal while the scattering from SH4UD is suppressed. In contrast, in 100% D2O solvent, the signal is dominated by SH4UD and the signal from SH3 and SH2 is suppressed.
[Reprinted with permission from Gurumoorthy et al. DOI:10.1021/acs.biomac.2c01158. Copyright 2023 American Chemical Society.]
The Science
Researchers at the Center for Structural Molecular Biology and the Center for Biophysics at Oak Ridge National Laboratory developed a segmental labeling procedure to study interactions between an intrinsically disordered N-terminal protein domain and folded domains in the multidomain protein c-Src kinase. The procedure, called domain-selective deuterium isotopic labeling or segmental labeling, was combined with small-angle neutron scattering (SANS). Together, these techniques enable functionally important disordered regions to be studied in multidomain proteins, providing structural insights that are difficult if not impossible to obtain with X-ray-based or electron-based structural characterization techniques.
The Impact
This study shows for the first time that a disordered region of c-Src kinase modulates the structure of the neighboring folded domain. The observation may have important implications for allosteric interactions with binding partners.
This type of structural information is difficult if not impossible to obtain with other structural characterization techniques using X-rays and electrons. The segmental labeling approach can be broadly applied to study functionally important disordered regions in other multidomain proteins involved in cell signaling and other biological processes in biological and environmentally relevant systems.
The Summary
The protein c-Src kinase is a multidomain non-receptor tyrosine kinase associated with many types of cancer. Although the structural properties of the protein’s folded catalytic and regulatory domains (SH3-SH2) have been extensively characterized, less is known about the N-terminal disordered region (SH4UD).
Protiated SH4UD was enzymatically ligated to deuterated SH3-SH2 domains to synthesize a single polypeptide chain of (SH4UD)H-(SH3-SH2)D. Contrast variation SANS showed that in the presence of SH4UD, the radius of gyration (Rg) of SH3-SH2 increases, indicating that it has a more extended conformation. Hamiltonian replica exchange molecular dynamics simulations provide a detailed molecular description of the structural changes in SH4UD-SH3-SH2. The simulations showed that the regulatory loops of SH3 undergo significant conformational changes in the presence of SH4UD while SH2 remains largely unchanged.
Funding
This work is supported by the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle LLC and by project ERKPA14 funded by BER in the DOE Office of Science. The authors thank Dr. Kevin Weiss for technical support for bio-deuteration. V.G. acknowledges the support of University of Tennessee Knoxville Science Alliance agency for providing financial support through a Graduate Advancement & Training Education (GATE) fellowship. Neutron scattering experiments on Bio-SANS were supported by the Center for Structural Molecular Biology funded by DOE BER project ERKP291. Computational work was supported by the National Energy Research Scientific Computing Center (contract no. DE-AC02-05CH11231) and The Oak Ridge Leadership Computing Facility (contract no. DE-AC05-00OR22725). This research used resources at the High Flux Isotope Reactor and Spallation Neutron Source, a DOE Office of Science User Facility operated by the Oak Ridge National Laboratory.
Related Links
References
Gurumoorthy, V., et al. 2023. “Disordered Domain Shifts the Conformational Ensemble of the Folded Regulatory Domain of the Multidomain Oncoprotein c-Src,” Biomacromolecules 24(2), 714–23. DOI:10.1021/acs.biomac.2c01158
