Modulation of Plant Hormones by GH3 Proteins
05/24/2012
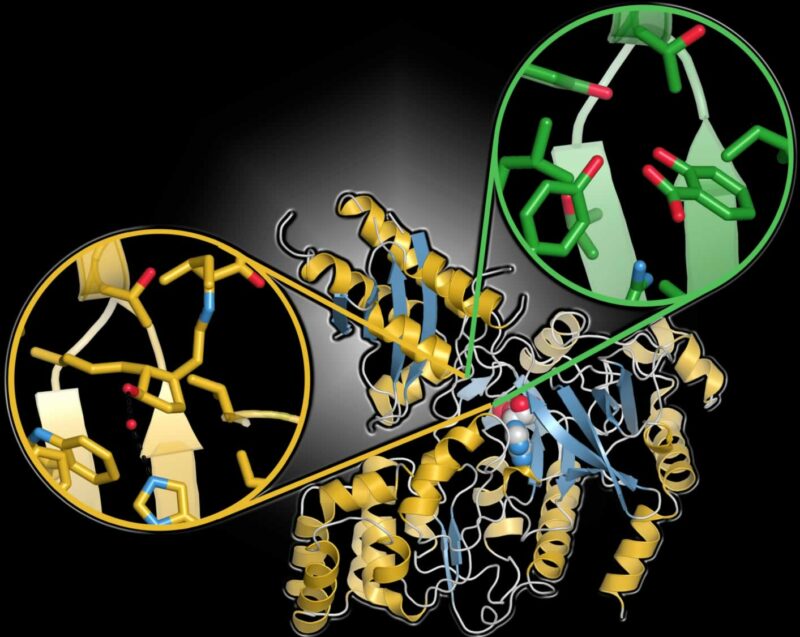
X-ray structures of various GH3 proteins, which modulate plant hormones, reveal a common three-dimensional structure (gold and blue ribbon diagram) but variability in the hormone binding site (two examples in circle insets). This figure illustrates the structural differences in the binding sites of two different GH3 proteins: the jasmonic acid binding site of Arabidopsis thaliana GH3.11/JAR1 (gold) and the salicylic acid binding site of A. thaliana GH3.12/PBS3 (green). [Courtesy of Argonne National Laboratory.]
Plants respond to developmental cues and environmental stresses by controlling the level and activity of various hormones. Within the auxin-responsive GH3 enzyme family, a highly adaptable protein scaffold enables the evolution of promiscuous activity and leads to the diversification of substrate specificity and the evolution of metabolic control systems.
Newly reported crystal structures for two acyl acid amido synthetase enzymes belonging to the GH3 family from Arabidopsis provide a glimpse into substrate recognition and the control of hormones involved in plant growth, development, and defense. The findings lend insight into the reaction chemistries that add functional diversity to hormone signaling pathways.
Funding Acknowledgements
The research was conducted using resources at the Advanced Photon Source at Argonne National Laboratory. Work supported by National Science Foundation (NSF) grant MCB-1157771 to J.M.J. C.S.W. supported by U.S. Department of Agriculture (USDA) National Institute of Food and Agriculture (NIFA) predoctoral fellowship (MOW-2010-05240), and J.H. supported by American Society of Plant Biologists (ASPB) Summer Undergraduate Research Fellowship award and the Howard Hughes Medical Institute (HHMI)–Washington University in St. Louis Summer Scholars Program in Biology and Biomedical Research. Portions of this research carried out at European Synchrotron Radiation Facility (ESRF) and Argonne National Laboratory (ANL) Structural Biology Center (SBC) of the Advanced Photon Source (APS), a national user facility operated by the University of Chicago for the Office of Biological and Environmental Research, U.S. Department of Energy (DOE) Office of Science (DE-AC02-06CH11357). Atomic coordinates and structure factors deposited in Protein Data Bank (PDB; accession codes noted in table S2).
Related Links
References
Westfall, C. S., et al. 2012. “Structural Basis for Prereceptor Modulation of Plant Hormones by GH3 Proteins,” Science 336, 1708–11. [DOI: 10.1126/science.1221863]
