Structure of a Flavoenzyme Assembly Intermediate
01/18/2018
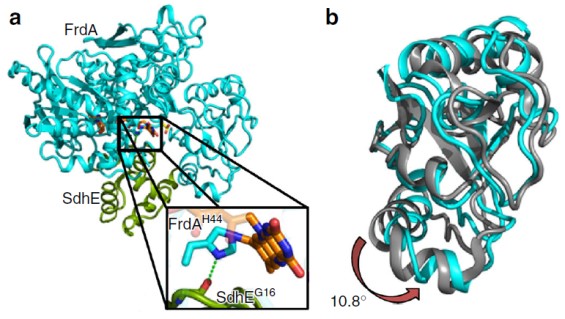
(a) The structure of the FrdA-SdhE flavoenzyme assembly intermediate: flavoprotein subunit FrdA (cyan), assembly factor SdhE (green), flavin adenine dinucleotide FAD (orange sticks), and malonate (yellow sticks). The boxed region highlights the covalent interaction between the FAD and the enzyme. (b) Overlay of the flavin-binding domains of the FrdA subunit from the FrdA-SdhE intermediate (cyan) and the FrdA subunit from the mature assembled FrdABCD complex (gray). A rotation of 10.8° is observed in the capping domain of the assembly intermediate when compared to assembled FrdABCD. [From Sharma, P., et al. “Crystal structure of an assembly intermediate of respiratory Complex II,” Nat. Commun. 9, 274 (2018). DOI:10.1038/s41467-017-02713-8. Reused under a Creative Commons license (CC BY 4.0, https://creativecommons.org/licenses/by/4.0/.)]
The Summary
Enzymes frequently depend on an electron transport cofactor for executing catalytic functions such as reduction-oxidation (redox) reactions. For flavoenzymes, the cofactor is flavin adenine dinucleotide (FAD), whose binding type with the enzyme impacts the redox potential and thus reaction chemistry, such as for metabolism and detoxification.
Researchers discovered that the structure of an assembled flavoenzyme intermediate reveals the mechanism of covalent flavin binding in respiration. Assembly factors include SdhAF2 in humans, SdhE in Escherichia coli, and Sdh5 in yeast. Other revelations include that mitochondrial flavoenzymes drive both noncovalent and covalent redox reactions and that the assembly factor (SdhE, a small protein of ~90 to 140 amino acids, conserved in all kingdoms) in the structure of the SdhE:FrdA complex with covalent FAD stabilizes a conformation of the flavoprotein subunit FrdA that favors succinate oxidation.
Researchers fixed the E. coli FrdA-SdhE intermediate via site-specific crosslinking, resolving the structure to 2.6 angstroms (Å). This study identified that SdhE stabilizes an FrdA conformation that likely enables the mechanism of autocatalytic covalent flavinylation. FrdA’s FAD-binding domain and capping domain both interact with SdhE, but structural data revealed a 10.8° difference in their angles.
The investigators believe that domain rotation affects flavinylation, showing that enzymes are tuned to catalyze reactions in different ways and that conformational diversity can directly relate to catalytic mechanism diversity.
Funding
Supported by Department of Veterans Affairs (DVA; BX001077 to G.C.) and National Institutes of Health (NIH; GM061606 to G.C. and T.M.I.). G.C. (recipient of a Senior Research Career Scientist award, #IK6B004215 from DVA). Vanderbilt University crystallization facility support: S10 RR026915. Use of Stanford Synchrotron Radiation Lightsource (SSRL), SLAC National Accelerator Laboratory (SLAC), supported by Office of Basic Energy Sciences (OBES), U.S. Department of Energy (DOE) Office of Science, under Contract No. DE-AC02-76SF00515. SSRL Structural Molecular Biology Program supported by Office of Biological and Environmental Research (OBER), DOE Office of Science, and by the NIH National Institute of General Medical Sciences (NIGMS; including P41GM103393).
Related Links
References
Sharma, P., et al. “Crystal structure of an assembly intermediate of respiratory Complex II.” Nat. Commun. 9, 274 (2018). DOI:10.1038/s41467-017-02713-8.
