Targeting a Critical Molecular Switch in COVID-19
07/31/2020
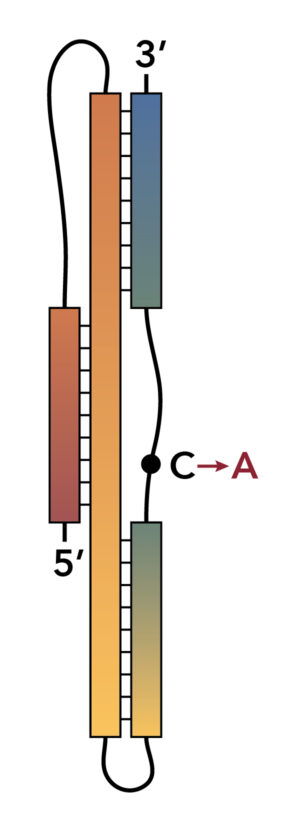
Solution X-ray scattering (SAXS) helps reveal a potential molecular target that may impair SARS-CoV-2 replication. The image represents a particular mRNA molecule from SARS-CoV, the virus that causes MERS, that controls the expression of viral proteins through a process called programmed −1 ribosomal frameshift (−1 PRF). The conformation and sequence are remarkably similar to that of SARS-CoV-2, in which a cytosine nucleotide is replaced by adenine (red). This suggests that compounds that inhibit -1 PRF in SARS-CoV may likewise inhibit SARS-CoV-2 replication. [Courtesy Brookhaven National Laboratory.]
The Science
Scientists studied the 3-D structure of parts of the COVID-19 virus and found a potential inhibitor for virus replication. They compared the three-stemmed RNA pseudoknots of SARS-CoV and SARS-CoV-2 that function as critical switches in the replication pathways of these viruses and found a small molecule that could inhibit their function.
The Impact
This work shows that such inhibitors may possibly be used to fight the COVID-19 pandemic.
Summary
The COVID-19 pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and researchers continue to work on treatments and vaccines, many of which can take years to develop. A good short-term strategy may lie in identifying virus-specific targets for small-molecule-based interventions.
In this work, a research team investigated how the virus replicates by studying a process called programmed −1 ribosomal frameshift (−1 PRF). In this process, a ribonucleic acid (RNA) pseudoknot acts as a critical switch in the replication programs of the virus and is therefore a promising drug target to inhibit virus replication.
The team used small-angle x-ray scattering (SAXS) analyses at the Life Science X-ray Scattering (LiX) beamline at the National Synchrotron Light Source II (NSLS-II) to compare the structure of the three-stemmed pseudoknots of SARS-CoV and SARS-CoV-2. SARS-CoV is a coronavirus strain that caused an epidemic 17 years ago and has been well studied since. NSLS-II is a U.S. Department of Energy Office of Science User facility located at DOE’s Brookhaven National Laboratory.
The team showed that the pseudoknots have a similar structure and function. This enabled testing whether a small molecule known to inhibit SARS-CoV frameshifting would be similarly effective against SARS-CoV-2. The results suggest that such frameshift inhibitors may be promising lead compounds to combat the COVID-19 pandemic.
Funding Acknowledgements
This research used resources including the LiX beamline of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The Center for BioMolecular Structure (CBMS) is primarily supported by the National Institutes of Health, National Institute of General Medical Sciences (NIGMS) through a Center Core P30 Grant (P30GM133893), and by the DOE Office of Biological and Environmental Research (KP1605010).
Related Links
- BER Resource: Center for BioMolecular Structure
- Feature Story: Targeting a Critical Molecular Switch in COVID-19
References
J. A. Kelly, A. N. Olson, K. Neupane, S. Munshi, J. San Emeterio, L. Pollack, M. T. Woodside, J. D. Dinman. “Structural and functional conservation of the programmed −1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2).” J. Biol. Chem. 295(31):10741-10748 (2020). [DOI: 10.1074/jbc.AC120.013449]
