The CRISPR Target-Recognition Mechanism
07/20/2017
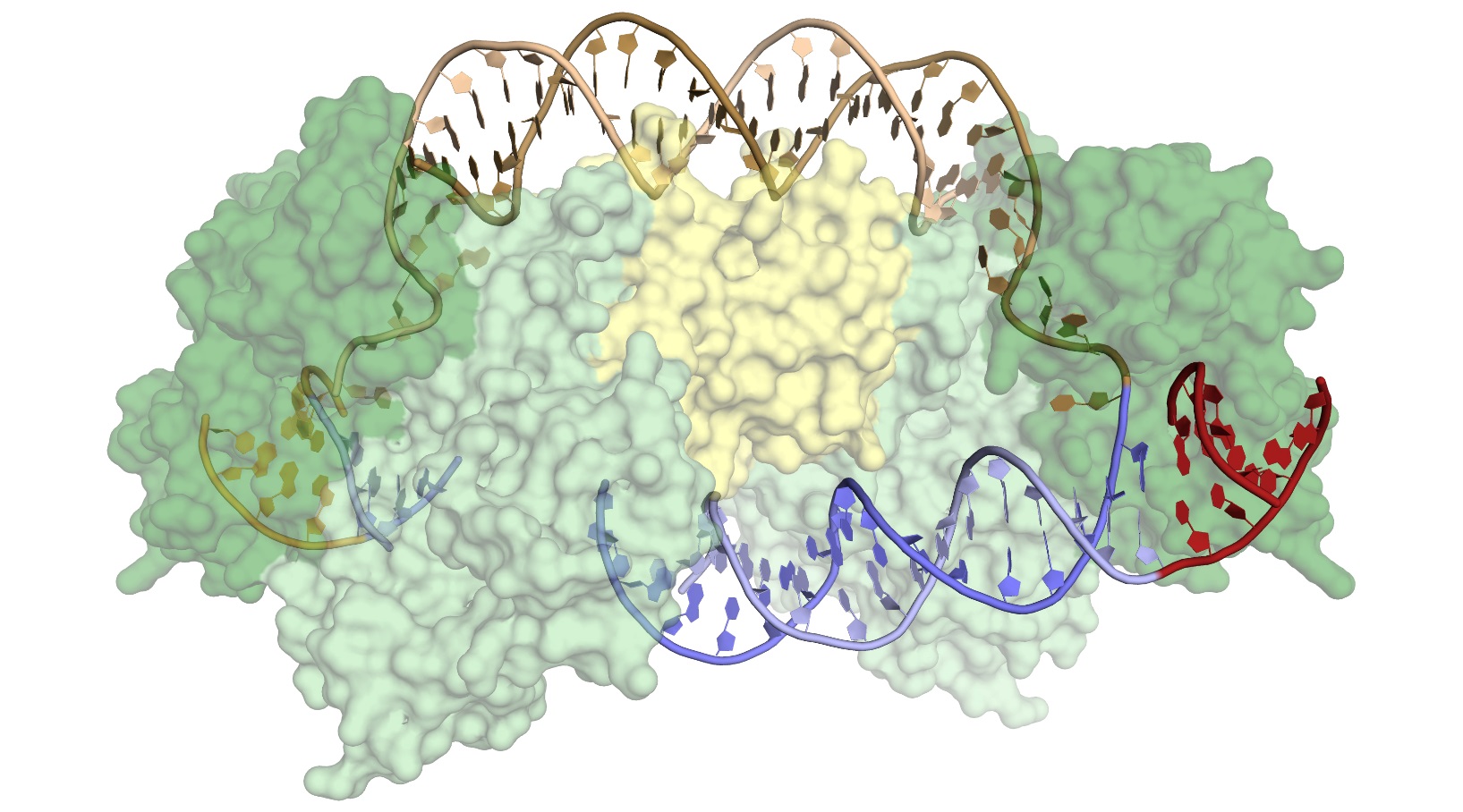
Surface representation of the Cas1-Cas2 complex, consisting of four Cas1 proteins (light and dark green) and two Cas2 proteins (yellow). Donor DNA (brown) is being integrated into the target DNA (blue), at a precise location in the CRISPR array, following a short leader sequence (red). [From Wright, A. V., et al. “Structures of the CRISPR Genome Integration Complex,” Science 357(6356), 1113–1118 (2017). [DOI:10.1126/science.aao0679. Reprinted with permission from AAAS*.] * Readers may view, browse, and/or download material for temporary copying purposes only, provided these uses are for noncommercial personal purposes. Except as provided by law, this material may not be further reproduced, distributed, transmitted, modified, adapted, performed, displayed, published, or sold in whole or in part, without prior written permission from the publisher.
Bacterial DNA is characterized by regions of clustered regularly interspaced short palindromic repeats (CRISPRs) and associated Cas proteins (CRISPR-associated endonucleases). The CRISPR-Cas system has revolutionized gene editing by vastly simplifying the insertion of short snippets of new (“donor”) DNA into very specific locations of target DNA.
Researchers in this study have discovered how Cas proteins recognize their target locations with such great specificity. They used x-ray crystallography to solve the structures of Cas1 and Cas2—responsible for DNA-snippet capture and integration—as the proteins were bound to synthesized DNA strands designed to mimic different stages of the process. The research also demonstrated how the system works in its native context as part of a bacterial immune system and how Cas proteins act as general-purpose molecular recording devices—tools for encoding information in genomes.
Cas1 appears to have evolved from a more “promiscuous” (less selective) type of enzyme that catalyzes the movement of DNA sequences from one position to another (a transposase). At some point, Cas1 acquired an unusual degree of specificity for a particular location in the bacterial genome, the CRISPR array. This specificity is critical to the bacteria, both for acquiring immunity and for avoiding genome damage caused by the insertion of viral fragments at the wrong location.
The researchers wanted to learn how Cas1-Cas2 proteins recognize the target sequence to enable comparison with previously studied transposases and integrases (i.e., enzymes that catalyze the integration of donor DNA into target DNA) and to determine whether the proteins can be altered to recognize new sequences for custom applications. To investigate this, they crystallized Cas1-Cas2 in complex with preformed DNA strands that mimicked reaction intermediates and products.
X-ray crystallography revealed that the structures showed substantial distortions in the target DNA, but there were surprisingly few sequence-specific contacts with the Cas1-Cas2 complex, and the DNA’s resulting flexibility produced disorder in the crystals. Attempts to model the DNA across the disordered sections showed that the DNA had to be even more distorted. Cryoelectron microscopy experiments, coupled with the crystallography data, confirmed that an accessor protein called the integration host factor (IHF) introduces an additional sharp bend in the DNA, bringing an upstream recognition sequence into contact with Cas1 to increase both the specificity and efficiency of integration.
The architecture of the CRISPR integration complex suggests that subtle adjustment of the distance between Cas1 active sites could reprogram the system to recognize different target sites. Changes in its architecture could be exploited, thereby, for genome tagging applications and also may explain the natural divergence of CRISPR arrays in bacteria.
Instruments and Facilities
X-ray macromolecular crystallography; beamline 8.3.1; protein crystallography (PX); and scattering/diffraction at the Advanced Light Source at Lawrence Berkeley National Laboratory; Stanford Synchrotron Radiation Light Source 9-2 beamline.
Funding Acknowledgements
Advanced Light Source (ALS) 8.3.1 beamline, Lawrence Berkeley National Laboratory (LBNL), and Stanford Synchrotron Radiation Lightsource (SSRL) 9-2 beamline, SLAC National Accelerator Laboratory (SLAC), for assistance with data collection. ALS Beamline 8.3.1, is operated by University of California Office of the President, Multicampus Research Programs and Initiatives (grant MR-15-328599), and Program for Breakthrough Biomedical Research, partially funded by the Sandler Foundation. Use of SSRL supported by the Office of Basic Energy Sciences (OBES), U.S. Department of Energy (DOE) Office of Science, under contract no. DE-AC02-76SF00515. Electron microscopy (EM) data collected in Howard Hughes Medical Institute (HHMI) EM facility located at University of California, Berkeley. SSRL Structural Molecular Biology Program supported by DOE Office of Biological and Environmental Research (OBER) and the National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS; including grant no. P41GM103393). Project funded by U.S. National Science Foundation (NSF) grant no. 1244557 (to J.A.D.) and NIGMS grant no. 1P50GM102706-01 (to J. H. Cate). A.V.W. and K.W.D. support: NSF Graduate Research Fellowship; G.J.K. funding: HHMI. J.A.D. and E.N.: HHMI investigators and members of the Center for RNA Systems Biology. Atomic coordinates and structure factors for the reported crystal structures deposited in the Protein Data Bank under accession codes 5VVJ (half-site–bound), 5VVK (pseudo–full-site–bound), and 5VVL (pseudo–full-site–bound with Ni2+). Cryo-EM structure and map deposited in the Protein Data Bank under accession code 5WFE and the Electron Microscopy Data Bank under accession code EMD-8827.
Related Links
- BER Resource: Structural Molecular Biology Resource
- Feature Story: The CRISPR Target-Recognition Mechanism
References
Wright, A. V., et al. “Structures of the CRISPR Genome Integration Complex,” Science 357(6356), 1113–1118 (2017). [DOI:10.1126/science.aao0679].
