Understanding How Bacteria Degrade N-Glycan
03/25/2021
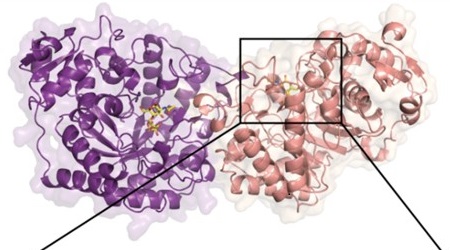
Crystal structure of a bacterial glycoside hydrolase enzyme showing two independent molecules in purple and pink. [Reprinted with permission from Higgins, et al. 2021. N-Glycan Degradation Pathways in Gut- and Soil-Dwelling Actinobacteria Share Common Core Genes. ACS Chem. Biol. 16 (4), 701-711. Copyright 2021 American Chemical Society.]
N-glycosylation is a fundamental protein modification found in both eukaryotes and archaea, but rarely bacteria. Despite lacking N-glycans, many commensal and pathogenic bacteria have developed mechanisms to degrade scavenged N-glycans for a variety of functions including nutrient acquisition; they express enzymes that cleave the glycan chain’s core. If the glycan is attached to a protein, it is first removed before cleavage. Although much is known about many enzymes responsible for N-glycan degradation, the enzymes involved in cleaving the N-glycan core have only recently been discovered. Thus, some of the structural details have yet to be characterized and little is known about their full distribution among bacterial strains.
Researchers used macromolecular X-ray crystallography to elucidate the structure of the active site of a family of glycoside hydrolases—enzymes responsible for glycan core degradation—from the soil bacterium Streptomyces cattleya and the gut bacterium Bifidobacterium longum. The study provides understanding of how these bacteria degrade glycan, pointing to an efficient mechanism for soil microbes to recycle decomposing biomass.
Related Links
References
Higgins, M.A.; Tegl, G.; MacDonald, S. S.; Arnal, G.; Brumer, H.; Withers, S. G.; Ryan, K. S. 2021. N-Glycan Degradation Pathways in Gut- and Soil-Dwelling Actinobacteria Share Common Core Genes. ACS Chem. Biol. 16 (4), 701-711. [DOI: 10.1021/acschembio.0c00995]
