Understanding Nitrogen Fixation in Bacteria
09/26/2014
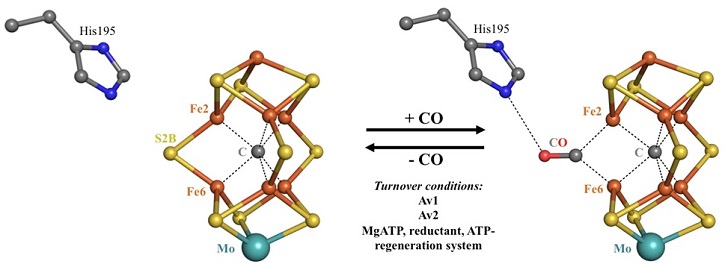
X-ray crystallography reveals reversible conformational changes occurring in a bacterial nitrogenase enzyme (the FeMo-cofactor active site is pictured here) during binding of carbon monoxide (CO), which simulates aspects of binding with the natural substrate dinitrogen (N2). [Image credit: Stanford Synchrotron Radiation Lightsource.]
Living organisms depend on nitrogen fixation to convert atmospheric nitrogen (N2) to a form they can incorporate into the basic building blocks of life, such as DNA and amino acids. Nitrogenase is the only known enzyme capable of fixing nitrogen. Therefore, understanding how nitrogenase performs its multi-electron reduction of N2 is highly important in ammonia fertilizer production, for energy efficiency (because industrial ammonia production consumes enormous amounts of energy), and for addressing climate change.
Scientists used X-ray macromolecular crystallography to determine the structure of the MoFe protein, one of two metalloproteins that comprise nitrogenase in the soil bacterium Azotobacter vinelandii, and how substrates bind to the enzyme’s active site. The structure and conformational changes that occur when bound to a substrate provide insight into how the N2 triple bond is reduced.
Funding Acknowledgements
Work supported by National Institutes of Health (NIH) grant GM45162 (D.C.R.), Deutsche Forschungsgemeinschaft grants EI-520/7 and RTG 1976, and European Research Council N-ABLE project (O.E.). Gordon and Betty Moore Foundation, Beckman Institute, and Sanofi-Aventis Bioengineering Research Program at Caltech: support of Molecular Observatory at Caltech and staff at Beamline 12–2, Stanford Synchrotron Radiation Lightsource (SSRL), for their assistance with data collection. SSRL is operated for the U.S. Department of Energy (DOE) Office of Science and supported by its Office of Biological and Environmental Research (OBER) and by the National Institutes of Health’s (NIH) National Institute of General Medical Sciences (NIGMS; P41GM103393) and National Center for Research Resources (NCRR; P41RR001209). Center for Environmental Microbial Interactions: support of microbiology research at Caltech. Coordinates and structure factors deposited in Protein Data Bank of the Research Collaboratory for Structural Bioinformatics, with IDs 4TKV (Av1-CO) and 4TKU (Av1 reactivated).
Related Links
- BER Resource: Structural Molecular Biology Resource
- Feature Story: Reversible CO-binding to the active site of nitrogenase
References
Spatzal, K.A. Perez, O. Einsle, J.B. Howard, D.C. Rees, “Ligand binding to the FeMo-cofactor: Structures of CO-bound and reactivated nitrogenase” Science 345, 1620-1623 (2014), doi: 10.1126/science.1256679.
