Using Neutrons to Resolve Plasma Membrane Organization in Live Bacteria
05/23/2017
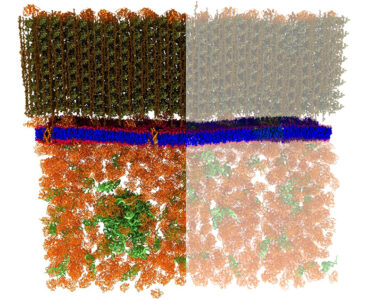
(Left) The cell wall (greenish brown, at top) of Bacillus subtilis, along with the bacterium’s plasma membrane (blue and red), and a portion of cytoplasm (rust, at bottom). (Right) SANS used in conjunction with selective hydrogen/deuterium labeling techniques revealed the structure of the plasma membrane, including nanoscale lipid domains, while blocking interfering signals from other cellular features. [From Nickels, J. D., et al. 2017. [DOI:10.1371/journal.pbio.2002214]. Reused under a Creative Commons license (CC BY 4.0, https://creativecommons.org/licenses/by/4.0/).]
The Summary
A new strategy devised by scientists at Oak Ridge National Laboratory used nondestructive small-angle neutron scattering (SANS) to reveal, for the first time, nanoscale membrane structures in living cells.
Neutron scattering spectra confirmed that the plasma membrane of the bacterium Bacillus subtilis is lamellar with an average hydrophobic thickness of 24 Ångstroms. The data also revealed that the membrane contains lipid features of approximately 40 nm or less in size, consistent with hypothesized “lipid rafts” in biological systems. The observation of lipid segregation in the plasma membrane of a bacterium is consistent with the notion of nanoscopic lipid assemblies, often described as lipid rafts in mammalian systems, implying that lipid domains are integral features of all biological membranes. Among their functions, lipid rafts are thought to play a vital role in cell signaling and facilitate movement of essential biomolecules in and out of the cell.
In addition to these scientific findings, the methods developed provide a new experimental platform for pursuing additional areas of inquiry (e.g., systematic in vivo investigations of cell membrane structure and response to diverse environmental stimuli). This new approach also may prove valuable, for example, in biomass feedstock and biofuel production, where bacterial cell membranes play important roles, and in biomedicine, where bacterial membrane domains affect antibiotic resistance. Furthermore, the strategy for “visualizing” the membrane can be used with other physical characterization techniques to examine additional cell structures such as the cell wall.
Instruments and Facilities
Small-angle neutron scattering (SANS) at the High Flux Isotope Reactor (HFIR) at Oak Ridge National Laboratory (ORNL). EQ-SANS at Spallation Neutron Source at ORNL. Oak Ridge Leadership Computing Facility at ORNL.
Funding
Work sponsored by Laboratory Directed Research and Development Program (grant number 6988) of Oak Ridge National Laboratory (ORNL), managed by UT-Battelle, LLC, for the U.S. Department of Energy (DOE) under Contract No. DE-AC05-00OR22725. Support for J.K.: Office of Basic Energy Sciences (OBES) Scientific User Facilities Division, DOE Office of Science. R.F.S.: Office of Biological and Environmental Research (OBER), DOE Office of Science (grant number ERKP-851). Resources of Oak Ridge Leadership Computing Facility at ORNL, supported by Facilities Division of DOE Office of Advanced Scientific Computing Research (OASCR). Small-angle neutron scattering (SANS) performed at ORNL using Bio-SANS instrument at the High Flux Isotope Reactor (HFIR), supported by DOE OBER’s Biological Systems Science Division, through ORNL Center for Structural Molecular Biology, and EQ-SANS instrument at Spallation Neutron Source (SNS) at ORNL, supported by the DOE OBES Scientific User Facilities Division (grant number ERKP-SNX).
Related Links
- BER Resource: Center for Structural Molecular Biology
- Feature Story: Neutrons provide the first nanoscale look at a living cell membrane
- Science Highlight
References
Heberle, F.A., et al. “Bilayer Thickness Mismatch Controls Domain Size in Model Membranes.” Journal of the American Chemical Society 135, 6853-6859 (2013). [DOI:10.1021/ja3113615]
Nickels, J.D. et al. “Lateral organization, bilayer asymmetry, and inter-leaflet coupling of biological membranes.” Chemistry and Physics of Lipids 192, 87-99 (2015). [DOI:10.1371/10.1016/j.chemphyslip.2015.07.012]
Nickels, J.D. et al. “Mechanical Properties of Nanoscopic Lipid Domains.” Journal of the American Chemical Society 137, 15772-15780 (2015). [DOI:10.1021/jacs.5b08894]
Nickels, J. D., et al. “The In Vivo Structure of Biological Membranes and Evidence for Lipid Domains.” PLOS Biology 15(5): e2002214 (2017). [DOI:10.1371/journal.pbio.2002214]
